Abstract
Key message
An unambiguous nomenclature is proposed for the twenty-eight-member LOB domain transcription factor family in Brachypodium . Expression analysis provides unique transcript patterns that are characteristic of a wide range of organs and plant parts.
Abstract
LOB (lateral organ boundaries)-domain proteins define a family of plant-specific transcription factors involved in developmental processes from embryogenesis to seed production. They play a crucial role in shaping the plant architecture through coordinating cell fate at meristem to organ boundaries. Despite their high potential importance, our knowledge of them is limited, especially in the case of monocots. In this study, we characterized LOB domain protein coding genes (LBDs) of Brachypodium distachyon, a model plant for grasses, and present their phylogenetic relationships and an overall spatial expression study. In the Brachypodium genome database, 28 LBDs were found and then classified based on the presence of highly conserved LOB domain motif. Their transcript amounts were measured via quantitative real-time RT-PCR in 37 different plant parts from root tip to generative organs. Comprehensive phylogenetic analysis suggests that there are neither Brachypodium- nor monocot-specific lineages among LBDs, but there are differences in terms of complexity of subclasses between monocots and dicots. Although LBDs in Brachypodium have wide variation of tissue-specific expression and relative transcript levels, overall expression patterns show similarity to their counterparts in other species. The varying transcript profiles we observed support the hypothesis that Brachypodium LBDs have diverse but conserved functions in plant organogenesis.





Similar content being viewed by others
References
Ariel FD, Diet A, Crespi M, Chan RL (2010) The LOB-like transcription factor Mt LBD1 controls Medicago truncatula root architecture under salt stress. Plant Signal Behav 5:1666–1668. doi:10.4161/psb.5.12.14020
Bell EM, W-c Lin, Husbands AY et al (2012) Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci USA 109:21146–21151. doi:10.1073/pnas.1210789109
Berckmans B, Vassileva V, Schmid SPC et al (2011) Auxin-Dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23:3671–3683. doi:10.1105/tpc.111.088377
Birnbaum KD, Alvarado AS (2008) Slicing across kingdoms: regeneration in plants and animals. Cell 132:697–710. doi:10.1016/j.cell.2008.01.040
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691. doi:10.1105/tpc.021410
Borghi L, Bureau M, Rüdiger S (2007) Arabidopsis JAGGED LATERAL ORGANS Is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19:1795–1808. doi:10.1105/tpc.106.047159
Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S (2006) ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18(3):574–585
Brutnell TP, Bennetzen JL, Vogel JP (2015) Brachypodium distachyon and Setaria viridis: model genetic systems for the grasses. Annu Rev Plant Biol 66:465–485. doi:10.1146/annurev-arplant-042811-105528
Bureau M, Rast M, Illmer J, Rüdiger S (2010) JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol Biol 74:479–491. doi:10.1007/s11103-010-9688-2
Cabrera J, Díaz-Manzano FE, Sanchez M et al (2014) A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol 203:632–645. doi:10.1111/nph.12826
Catalan P, Chalhoub B, Chochois V et al (2014) Update on the genomics and basic biology of Brachypodium. Trends Plant Sci 19:414–418. doi:10.1016/j.tplants.2014.05.002
Chanderbali AS, He F, Soltis PS, Soltis DE (2015) Out of the water: origin and diversification of the LBD gene family. Mol Biol Evol 32:1996–2000
Coudert Y, Dievart A, Droc G, Gantet P (2013) ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol Biol Evol 30:569–572
Coudert Y, Le VAT, Adam H et al (2015) Identification of CROWN ROOTLESS1-regulated genes in rice reveals specific and conserved elements of postembryonic root formation. New Phytol 206:243–254. doi:10.1111/nph.13196
Deng W, Wang Y, Liu Z, Cheng H, Xue Y (2014) HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi:10.1371/journal.pone.0111988
Draper J, Mur LAJ, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge APM (2001) Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol 127:1539–1555
Evans MMS (2007) The indeterminate gametophyte1 Gene of Maize Encodes a LOB Domain Protein Required for Embryo Sac and Leaf Development. Plant Cell 19:46–62. doi:10.1105/tpc.106.047506
Fan M, Xu C, Xu K, Hu Y (2012) LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22:1169–1180. doi:10.1038/cr.2012.63
Feng Z, Zhu J, Du X, Cui X (2012) Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta 236:1227–1237. doi:10.1007/s00425-012-1673-3
Girin T, David LC, Chardin C, Sibout R, Krapp A, Ferrario-Méry S, Daniel-Vedele F (2014) Brachypodium: a promising hub between model species and cereals. J Exp Bot 65:5683–5696
Goh T, Joi S, Mimura T, Fukaki H (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139:883–893
Guo M, Thomas J, Collins G, Timmermans MCP (2008) Direct repression of KNOX Loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20:48–58. doi:10.1105/tpc.107.056127
Hetz W, Hochholdinger F, Schwall M, Feix G (1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10:845–857. doi:10.1046/j.1365-313X.1996.10050845.x
Hong S-Y, Seo PJ, Yang M-S, Xiang F, Park C-M (2008) Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol 8:112. doi:10.1186/1471-2229-8-112
Hu Y, Zhang J, Jia H et al (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc Natl Acad Sci USA 111:E521–E529. doi:10.1073/pnas.1313271111
Husbands A, Bell EM, Shuai B, Smith HMS, Springer PS (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res 35:6663–6671. doi:10.1093/nar/gkm775
Inukai Y, Sakamoto T, Ueguchi-Tanaka M et al (2005) Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17:1387–1396. doi:10.1105/tpc.105.030981
Iwakawa H, Ueno Y, Semiarti E et al (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43:467–478
Jaakola L, Pirttilä A, Halonen M, Hohtola A (2001) Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol 19:201–203. doi:10.1385/mb:19:2:201
Kellogg EA (2015) Brachypodium distachyon as a genetic model system. Annu Rev Genet 49:1–20. doi:10.1146/annurev-genet-112414-055135
Kim M-J, Kim J (2012) Identification of nuclear localization signal in ASYMMETRIC LEAVES2-LIKE18/LATERAL ORGAN BOUNDARIES DOMAIN16 (ASL18/LBD16) from Arabidopsis. J Plant Physiol 169:1221–1226. doi:10.1016/j.jplph.2012.04.004
Landschulz WH, Johnson PF, McKnight SL (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759–1764. doi:10.1126/science.3289117
Lee HW, Kim NY, Lee DJ, Kim J (2009) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol 151:1377–1389
Lee T-H, Tang H, Wang X, Paterson AH (2013) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 41:D1152–D1158. doi:10.1093/nar/gks1104
Li A, Zhang Y, Wu X, Tang W, Wu R, Dai Z et al (2008) DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol Biol 66(5):491–502
Li C, Zou X, Zhang C, Shao Q, Liu J, Liu B et al (2016) OsLBD3-7 overexpression induced adaxially rolled leaves in rice. PLoS One 11(6):e0156413
Liu H, Wang S, Yu X et al (2005) ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43:47–56. doi:10.1111/j.1365-313X.2005.02434.x
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Ma Y, Wang F, Guo J, Zhang X (2009) Rice OsAS2 gene, a member of LOB domain family, functions in the regulation of shoot differentiation and leaf development. J Plant Biol 52:374–381. doi:10.1007/s12374-009-9048-4
Majer C, Hochholdinger F (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci 16:47–52. doi:10.1016/j.tplants.2010.09.009
Matsumura Y, Iwakawa H, Machida Y, Machida C (2009) Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J 58:525–537. doi:10.1111/j.1365-313X.2009.03797.x
Naito T, Yamashino T, Kiba T, Koizumi N, Kojima M, Sakakibara H, Mizuno T (2007) A Link between Cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci Biotechnol Biochem 71:1269–1278. doi:10.1271/bbb.60681
Oh SA, Park KS, Twell D, Park SK (2010) The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J 64:839–850. doi:10.1111/j.1365-313X.2010.04374.x
Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19:118–130
Opanowicz M, Vain P, Draper J, Parker D, Doonan JH (2008) Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci 13:172–177. doi:10.1016/j.tplants.2008.01.007
Pennington RT, Willis KJ, McElwain JC (2002) The evolution of plants. Ann Bot 90:678–679
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Phelps-Durr TL, Thomas J, Vahab P, Timmermans MCP (2005) Maize rough sheath2 and Its Arabidopsis Orthologue ASYMMETRIC LEAVES1 Interact with HIRA, a predicted Histone Chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17:2886–2898. doi:10.1105/tpc.105.035477
Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21:3718–3731. doi:10.1105/tpc.109.071506
Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K (2015) PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res 43:D974–D981. doi:10.1093/nar/gku986
Rast MI, Rüdiger S (2008) The meristem-to-organ boundary: more than an extremity of anything. Curr Opin Genet Dev 18:287–294. doi:10.1016/j.gde.2008.05.005
Rast MI, Rüdiger S (2012) Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 24:2917–2933. doi:10.1105/tpc.112.099978
Rubin G, Tohge T, Matsuda F, Saito K, Scheible W-R (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21:3567–3584. doi:10.1105/tpc.109.067041
Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128:1771–1783
Shuai B, Reynaga-Peña CG, Springer PS (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol 129:747–761
Soyano T, Thitamadee S, Machida Y, Chua N-H (2008) ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20:3359–3373. doi:10.1105/tpc.108.061796
Sun X, Feng Z, Meng L, Zhu J, Geitmann A (2013) Arabidopsis ASL11/LBD15 is involved in shoot apical meristem development and regulates WUS expression. Planta 237(5):1367–1378
Svec D, Tichopad A, Novosadova V, Pfaffl MW, Kubista M (2015) How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantif 3:9–16. doi:10.1016/j.bdq.2015.01.005
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Taramino G, Sauer M, Stauffer JL, Multani D, Niu X, Sakai H, Hochholdinger F (2007) The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J 50:649–659. doi:10.1111/j.1365-313X.2007.03075.x
Thatcher LF, Powell JJ, Aitken EAB, Kazan K, Manners JM (2012) The lateral organ boundaries domain transcription factor LBD20 functions in fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol 160:407–418. doi:10.1104/pp.112.199067
Vogel J, Bragg J (2009) Brachypodium distachyon, a new model for the Triticeae. In: Muehlbauer GJ, Feuillet C (eds) Genetics and genomics of the Triticeae, vol 7. Plant genetics and genomics: crops and models. Springer, USA, pp 427–449. doi:10.1007/978-0-387-77489-3_16
Vogel J, Garvin D, Mockler T, Schmutz J, Rokhsar D, Bevan M (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463:763–768. http://www.nature.com/nature/journal/v463/n7282/suppinfo/nature08747_S1.html
Wang X, Zhang S, Su L, Liu X, Hao Y (2013) A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES Domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS One 8:e57044. doi:10.1371/journal.pone.0057044
W-c Lin, Shuai B, Springer PS (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES—domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15:2241–2252. doi:10.1105/tpc.014969
Xu B, Li Z, Zhu Y, Wang H, Ma H, Dong A, Huang H (2008) Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol 146:566–575. doi:10.1104/pp.107.113787
Xu C, Luo F, Hochholdinger F (2016) LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci 21(2):159–167
Yang Y, Yu X, Wu P (2006) Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol 39:248–262. doi:10.1016/j.ympev.2005.09.016
You J, Zhang L, Song B, Qi X, Chan Z (2015) Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon. PLoS One 10:e0122027. doi:10.1371/journal.pone.0122027
Zentella R, Zhang Z-L, Park M et al (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19:3037–3057. doi:10.1105/tpc.107.054999
Zhang Y-M, Zhang S-Z, Zheng C-C (2014) Genomewide analysis of LATERAL ORGAN BOUNDARIES domain gene family in Zea mays. J Genetics 93:79–91. doi:10.1007/s12041-014-0342-7
Acknowledgments
This research was supported by the Hungarian Scientific Research Foundation (OTKA-K76273 and OTKA-K109719) and the Hungarian Governmental Grant GINOP-2.3.2-15-2016-00001. The authors are grateful to Judit Györgyey Ries and John Ries for their correction of the English of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by P. Puigdomenech.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2016_2057_MOESM1_ESM.tif
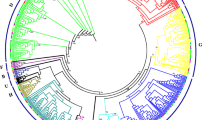
Online Resource 1 Combined phylogenetic tree of LOB-domain proteins from rice (Oryza sativa sp. Japonica) (medium blue dots), maize (Zea mays) (dark blue dots), Arabidopsis thaliana (red dots) and Brachypodium distachyon (light blue dots). The unrooted NJ tree was constructed with MEGA6 software based on amino acid sequences of conserved domain motif of 28 B. distachyon, 43 A. thaliana, 35 O. sativa and 44 maize LOB-domain proteins. The numbers on the branches are local bootstrap values calculated with 1000 replicates. Subfamilies are indicated by colour arches (TIFF 31431 kb)
299_2016_2057_MOESM2_ESM.rtf
Online Resource 2 Multiple sequence alignment of amino acid sequences of conserved domain motif of LOB-domain proteins from O. sativa, Zea mays, A. thaliana and B. distachyon. Invariant residues are marked with *. Consensus amino acid residues conserved in more than 50 % of proteins are indicated by grey background, while residues conserved in more than 75 % of proteins are indicated with withe font colour on black background. Os01g39040-A and Os01g39040-B highlighted with red font colour are two different LOB-domain motifs of the same protein, but presented in this alignment separately. The numbers mark the starting and ending position of LOB-domain motifs in each protein (RTF 1213 kb)
299_2016_2057_MOESM3_ESM.tif
Online Resource 3 The relative transcript amount of all 28 LBD genes of B. distachyon in root tip (R1), root differentiation zone (R3), 2nd leaf blade (L2), 3rd stem node (N3), flowers 1–2 days before pollination (F1), seeds 10 days after pollination (E3) and anthers. Root tips, 2nd leaf blades and 3rd stem nodes were collected from 14 DAG (days after germination) old plantlets; flowers 1–2 days before pollination and anthers were collected from 28 DAG old plantlets, seeds 10 days after pollination (10 DAP) were harvested from 40–45 DAG old plants grown under normal condition. The levels of gene expression were determined by quantitative real-time RT-PCR; at first relative to average transcript amount of EF1-α and UBC18 genes and second by relative to average transcript amount of each LBD genes. Means ±SD are shown (n = 2). Diagrams marked with black star are parts of Figure 5. Corresponding numeric data are listed in Online Resource 6 (TIFF 3727 kb)
299_2016_2057_MOESM4_ESM.xls
Online Resource 4 List of primer sequences used for quantitative real-time RT-PCR measurements. Some supporting information (genome position of BdLBD genes; length, weight and isoelectronic points of encoded proteins) are also represented (XLS 31 kb)
299_2016_2057_MOESM5_ESM.xlsx
Online Resource 5 Log10-transformed relative transcript amount data corresponding to expression profile analysis of 28 BdLBD genes in 37 different plant parts. Presented data is from one biological replicate derived from approximately 200 plants (XLSX 26 kb)
299_2016_2057_MOESM6_ESM.xlsx
Online Resource 6 Relative transcript amount data corresponding to biological replication (n = 2) of the expression pattern of 28 BdLBD genes in seven selected samples (root tip (R1), root differentiation zone (R3), 2nd leaf blade (L2), 3rd stem node (N3), flowers 1–2 days before pollination (F1), seeds 10 days after pollination (E3) and anthers) (XLSX 25 kb)
Rights and permissions
About this article
Cite this article
Gombos, M., Zombori, Z., Szécsényi, M. et al. Characterization of the LBD gene family in Brachypodium: a phylogenetic and transcriptional study. Plant Cell Rep 36, 61–79 (2017). https://doi.org/10.1007/s00299-016-2057-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-016-2057-0