Abstract
Key message
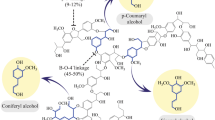
Two 2-phenylethanol biosynthetic pathways were constructed into Arabidopsis ; 2-phenylethanol biosynthesis led to reduced rate of lignin biosynthesis and increased cellulose-to-glucose conversion in the transgenic plants.
Abstract
Lignin is the second most abundant biopolymer on the planet with importance for various agro-industrial activities. The presence of lignin in cell walls, however, impedes biofuel production from lignocellulosic biomass. The phenylpropanoid pathway is responsible for the biosynthesis of lignin and other phenolic metabolites such as 2-phenylethanol. As one of the most used fragrance chemicals, 2-phenylethanol is synthesized in plants from l-phenylalanine which is the first specific intermediate towards lignin biosynthesis. Thus, it is interesting to prove the concept that the phenylpropanoid pathway can be modulated for reduction of lignin as well as production of natural value-added compounds. Here we conferred two 2-phenylethanol biosynthetic pathways constructed from plants and Saccharomyces cerevisiae into Arabidopsis. As anticipated, 2-phenylethanol was accumulated in transgenic plants. Moreover, the transformants showed 12–14 % reduction in lignin content and 9–13 % increase in cellulose content. Consequently, the glucose yield from cell wall hydrolysis was increased from 37.4 % in wild type to 49.9–52.1 % in transgenic plants with hot water pretreatment. The transgenic plants had normal development and even enhanced growth relative to the wild type. Our results indicate that the shunt of l-phenylalanine flux to the artificially constructed 2-phenylethanol biosynthetic pathway most likely reduced the rate of lignin biosynthesis in Arabidopsis.






Similar content being viewed by others
References
Abdel-Ghany SE, Day I, Heuberger AL, Broeckling CD, Reddy AS (2013) Metabolic engineering of Arabidopsis for butanetriol production using bacterial genes. Metab Eng 20:109–120
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54:519–546
Bonawitz ND, Chapple C (2013) Can genetic engineering of lignin biosynthesis be accomplished without an unacceptable yield penalty? Curr Opin Biotechnol 24:336–343
Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, Chapple C (2014) Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509(7500):376–380
Börnke F, Broer I (2010) Tailoring plant metabolism for the production of novel polymers and platform chemicals. Curr Opin Plant Biol 13(3):354–362
Campbell MM, Sederoff RR (1996) Variation in lignin content and composition. Mechanisms of control and implications for the genetic improvement of plants. Plant Physiol 110:3–13
Chai G, Wang Z, Tang X, Yu L, Qi G, Wang D, Yan X, Kong Y, Zhou G (2014) R2R3-MYB gene pairs in Populus: evolution and contribution to secondary wall formation and flowering time. J Exp Bot 65(15):4255–4269
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25:759–761
Douglas CJ (1996) Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci 1:171–178
Eudes A, George A, Mukerjee P, Kim JS, Pollet B, Benke PI, Yang F, Mitra P, Sun L, Cetinkol OP, Chabout S, Mouille G, Soubigou-Taconnat L, Balzergue S, Singh S, Holmes BM, Mukhopadhyay A, Keasling JD, Simmons BA, Lapierre C, Ralph J, Loqué D (2012) Biosynthesis and incorporation of side-chain-truncated lignin monomers to reduce lignin polymerization and enhance saccharification. Plant Biotechnol J 10(5):609–620
Eudes A, Liang Y, Mitra P, Loqué D (2014) Lignin bioengineering. Curr Opin Biotechnol 26:189–198
Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) part II: carbohydrates. J Vis Exp (37)
Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M Jr, Chen F, Foston M, Ragauskas A, Bouton J, Dixon RA, Wang ZY (2011a) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108(9):3803–3808
Fu C, Xiao X, Xi Y, Ge Y, Chen F, Bouton J, Dixon RA, Wang ZY (2011b) Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. Bioenerg Res 4(3):153–164
Gleba Y, Klimyuk V, Marillonnet S (2007) Viral vectors for the expression of proteins in plants. Curr Opin Biotechnol 18:134–141
Guo D, Chen F, Inoue K, Blount JW, Dixon RA (2001a) Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13:73–88
Guo D, Chen F, Wheeler J, Winder J, Selman S, Peterson M, Dixon RA (2001b) Improvement of in-rumen digestibility of alfalfa forage by genetic manipulation of lignin O-methyltransferases. Transgenic Res 10:457–464
Hazelwood LA, Daran JM, van Maris AJ, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74(8):2259–2266
Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, Tsai CJ, Chiang VL (1999) Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol 17(8):808–812
Jouanin L, Goujon T, de Nadaï V, Martin MT, Mila I, Vallet C, Pollet B, Yoshinaga A, Chabbert B, Petit-Conil M, Lapierre C (2000) Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol 123:1363–1374
Kaminaga Y, Schnepp J, Peel G, Kish CM, Ben-Nissan G, Weiss D, Orlova I, Lavie O, Rhodes D, Wood K, Porterfield DM, Cooper AJ, Schloss JV, Pichersky E, Vainstein A, Dudareva N (2006) Plant phenylacetaldehyde synthase is a bifunctional homotetrameric enzyme that catalyzes phenylalanine decarboxylation and oxidation. J Biol Chem 281(33):23357–23366
Karimi M, Bleys A, Vanderhaeghen R, Hilson P (2007) Building blocks for plant gene assembly. Plant Physiol 145(4):1183–1191
Li X, Weng JK, Chapple C (2008) Improvement of biomass through lignin modification. Plant J 54(4):569–581
Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, Pauly M, Cheng Z, Zhou Y (2009) Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J 60(6):1055–1069
Li X, Ximenes E, Kim Y, Slininger M, Meilan R, Ladisch M, Chapple C (2010) Lignin monomer composition affects Arabidopsis cell-wall degradability after liquid hot water pretreatment. Biotechnol Biofuels 3:27
Louie GV, Bowman ME, Tu Y, Mouradov A, Spangenberg G, Noel JP (2010) Structure-function analyses of a caffeic acid O-methyltransferase from perennial ryegrass reveal the molecular basis for substrate preference. Plant Cell 22:4114–4127
Pomar F, Merino F, Barceló AR (2002) O-4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220(1–2):17–28
Qi G, Hu R, Yu L, Chai G, Cao Y, Zuo R, Kong Y, Zhou G (2013) Two poplar cellulose synthase-like D genes, PdCSLD5 and PdCSLD6, are functionally conserved with Arabidopsis CSLD3. J Plant Physiol 170(14):1267–1276
Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344(6185):1246843
Reddy MS, Chen F, Shadle G, Jackson L, Aljoe H, Dixon RA (2005) Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc Natl Acad Sci USA 102:16573–16578
Rusanov K, Kovacheva N, Vosman B, Zhang L, Rajapakse S, Atanassov A, Atanassov I (2005) Microsatellite analysis of Rosa damascena Mill. accessions reveals genetic similarity between genotypes used for rose oil production and old Damask rose varieties. Theor Appl Genet 111(4):804–809
Sakai M, Hirata H, Sayama H, Sekiguchi K, Itano H, Asai T, Dohra H, Hara M, Watanabe N (2007) Production of 2-phenylethanol in roses as the dominant floral scent compound from l-phenylalanine by two key enzymes, a PLP-dependent decarboxylase and a phenylacetaldehyde reductase. Biosci Biotechnol Biochem 71(10):2408–2419
Samuel R, Pu Y, Jiang N, Fu C, Wang ZY, Ragauskas A (2014) Structural characterization of lignin in wild-type versus COMT down-regulated switchgrass. Front Energy Res 1:1–9
Sarath G, Mitchell RB, Sattler SE, Funnell D, Pedersen JF, Graybosch RA, Vogel KP (2008) Opportunities and roadblocks in utilizing forages and small grains for liquid fuels. J Ind Microbiol Biotechnol 35:343–354
Sewalt VJH, Ni W, Blount JW, Jung HG, Masoud SA, Howles PA, Lamb C, Dixon RA (1997a) Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of l-phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol 115:41–50
Sewalt VJH, Ni WT, Jung HG, Dixon RA (1997b) Lignin impact on fiber degradation: increased enzymatic digestibility of genetically engineered tobacco (Nicotiana tabacum) stems reduced in lignin content. J Agric Food Chem 45:1977–1983
Shen B, Li C, Tarczynski MC (2002) High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-l-methionine synthetase 3 gene. Plant J 29(3):371–380
Shen H, He X, Poovaiah CR, Wuddineh WA, Ma J, Mann DG, Wang H, Jackson L, Tang Y, Stewart CN Jr, Chen F, Dixon RA (2012) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193(1):121–136
Tang HM, Liu S, Hill-Skinner S, Wu W, Reed D, Yeh CT, Nettleton D, Schnable PS (2014) The maize brown midrib2 (bm2) gene encodes a methylenetetrahydrofolate reductase that contributes to lignin accumulation. Plant J 77(3):380–392
Tieman DM, Loucas HM, Kim JY, Clark DG, Klee HJ (2007) Tomato phenylacetaldehyde reductases catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochemistry 68(21):2660–2669
Tu Y, Rochfort S, Liu Z, Ran Y, Griffith M, Badenhorst P, Louie GV, Bowman ME, Smith KF, Noel JP, Mouradov A, Spangenberg G (2010) Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 22:3357–3373
Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6(1):46
Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010) Lignin biosynthesis and structure. Plant Physiol 153(3):895–905
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3(1):2–20
Ximenes E, Kim Y, Mosier N, Dien B, Ladisch M (2010) Inhibition of cellulases by phenols. Enzyme Microb Tech 46:170–176
Yu L, Zhang X, Li SS, Liu XY, Sun L, Liu HB, Iteku J, Zhou YF, Tai GH (2010) Rhamnogalacturonan I domains from ginseng pectin. Carbohydr Polym 79(4):811–817
Zhang K, Bhuiya MW, Pazo JR, Miao Y, Kim H, Ralph J, Liu CJ (2012) An engineered monolignol 4-o-methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell 24(7):3135–3152
Zhao Q, Tobimatsu Y, Zhou R, Pattathil S, Gallego-Giraldo L, Fu C, Jackson LA, Hahn MG, Kim H, Chen F, Ralph J, Dixon RA (2013) Loss of function of cinnamyl alcohol dehydrogenase 1 leads to unconventional lignin and a temperature-sensitive growth defect in Medicago truncatula. Proc Natl Acad Sci USA 110(33):13660–13665
Acknowledgments
This study was supported by the National Science and Technology Support Program (2013BAD22B01), the National Basic Research Program of China (2012CB114501), the National Natural Science Foundation of China (No. 31300502 and No. 31470291), China Postdoctoral Science Foundation, and the Youth Talent Plan of Chinese Academy of Agricultural Sciences to Y.Z.K.
Conflict of interest
We declare that we have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Q. Zhao.
G. Qi and D. Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Qi, G., Wang, D., Yu, L. et al. Metabolic engineering of 2-phenylethanol pathway producing fragrance chemical and reducing lignin in Arabidopsis . Plant Cell Rep 34, 1331–1342 (2015). https://doi.org/10.1007/s00299-015-1790-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1790-0