Abstract
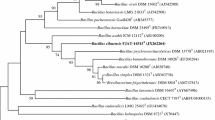
A Gram-staining-positive, motile, rod-shaped, aerobic bacterial strain FJAT-25496 T was isolated from a forest soil sample collected from Dafu County, Chengdu City, Sichuan Province, China. Strain FJAT-25496 T grew at 15–50 °C (optimum 35 °C), pH 6.0–10.0 (optimum 8.0), and NaCl tolerance of 0–4.0% (w/v) (optimum 0%). The strain contained iso-C15:0 and anteiso-C15:0 as the predominant cellular fatty acids as well as MK-7 as the only menaquinone. The major polar lipids were diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), and phosphatidyl glycerol (PG). Phylogenetic analyses based on 16S rRNA gene sequences showed that strain FJAT-25496 T was a member of the genus Bacillus, and most closely related to Bacillus solani FJAT-18043 T (97.8%), Bacillus praedii FJAT-25547 T (97.8%), Bacillus depressus BZ1T (97.7%), Bacillus oceanisediminis H2T (97.7%), and Bacillus ciccensis 5L6T (97.6%). Based on complete genome comparisons between strain FJAT-25496 T and the type strains of its closely related species, the average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values were 73.4–76.3% and 21.7–23.2%, respectively. The genomic DNA G + C content of strain FJAT-25496 T was 36.9%. On the basis of phenotypic, phylogenetic, chemotaxonomic, and genomic features, strain FJAT-25496 T is considered to represent a novel species of the genus Bacillus, for which the name Bacillus dafuensis sp. nov. is proposed. The type strain is FJAT-25496 T (= CCTCC AB 2019182 T = KCTC 43120 T).

Similar content being viewed by others
Abbreviations
- ANI:
-
Average Nucleotide Identity
- DPG:
-
Diphosphatidyl glycerol
- PME:
-
Phosphatidyl methyl ethanolamine
- PG:
-
Phosphatidyl glycerol
- PC:
-
Phosphatidyl choline
- UAPL:
-
Unidentified aminophospholipids
References
Cohn F (1872) Untrersuchungen über Bakterien Beitr Biol Pflanz 1:127–244
Liu B, Tao TS, Wang JP, Liu GH, Xiao RF, Chen MC (2016) Taxonomy of Bacillus. Science press, Beijing, China
Zhang L, Wu GL, Wang Y, Dai J, Fang CX (2011) Bacillus deserti sp. nov., a novel bacterium isolated from the desert of Xinjiang. China Antonie Van Leeuwenhoek 99:221–229
Romano I, Lama L, Nicolaus B, Gambacorta A, Giordano A (2005) Bacillus saliphilus sp. nov., isolated from a mineral pool in campania. Italy Int J Syst Evol Microbiol 55:159–163
Sultanpuram VR, Mothe T, Chintalapati S, Chintalapati VR (2017) Bacillus catenulatus sp. nov., an alkalitolerant bacterium isolated from a soda lake. Arch microbiol 199:1391–1397
Ma K, Chen X, Guo X, Wang H, Zhou S, Song J, Kong D, Zhu J, Dong W, He M, Hu G, Zhao B, Ruan Z (2016) Bacillus vini sp. nov. isolated from alcohol fermentation pit mud. Arch Microbiol 198:559–564
Tang YW, Ellis NM, Hopkins MK, Smith DH, Dodge DE, Persing DH (1998) Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J Clin Microbiol 36:3674–3679
Cihan AC, Tekin N, Ozcan B, Cokmus C (2012) The genetic diversity of genus Bacillus and the related genera revealed by 16S rRNA gene sequences and ardra analyses isolated from geothermal regions of turkey. Braz J Microbiol 43:309–324
Oliveira MGDM, Abels S, Zbinden R, Bloemberg GV, Zbinden A (2013) Accurate identification of fastidious Gram-negative rods: integration of both conventional phenotypic methods and 16S rRNA gene analysis. BMC Microbiol 13.
Nagata R, Takaki Y, Tame A, Nunoura T, Muto H, Mino S, Sawayama S, Takai K, Nakagawa S (2017) Lebetimonas natsushimae sp. nov., a novel strictly anaerobic, moderately thermophilic chemoautotroph isolated from a deep-sea hydrothermal vent polychaete nest in the Mid-Okinawa trough. Syst Appl Microbiol 40:352–356
Seck EH, Beye M, Traore SI, Khelaifia S, Michelle C, Couderc C, Brah S, Fournier PE, Raoult D, Bittar F (2017) Bacillus kwashiorkori sp. Nov., a new bacterial species isolated from a malnourished child using culturomics. MicrobiologyOpen 7(1):e00535
Togo AH, Awa D, Matthieu M, Lagier JC, Robert C, Pinto FD, Raout D, Fournier PE, Bittar F (2018) Draft genome and description of Eisenbergiella massiliensis strain AT11T: A new species isolated from human feces after bariatric surgery. Curr Microbiol 75:1274–1281
Dong X, Cai M (2001) Manual of systematic and determinative bacteriology. Science press, Beijing, China
Atlas RM (1993) Handbook of Microbiological Media. CRC Press, Boca Raton, Florida, USA
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society Microbiology, Washington, DC, pp 607–654
Chen YG, Cui XL, Pukall R, Li HM, Yang YL, Xu LH, Wen ML, Peng Q, Jiang CL (2007) Salinicoccus kunmingensis sp. nov., a moderately halophilic bacterium isolated from a salt mine in Yunnan, south-west China. Int J Syst Evol Microbiol 57:2327–2332
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC News Lett 20:1–6
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Schleifer KH (1985) Analysis of the chemical composition and primary structure of murein. Method Microbiol 18:123–156
MinnikinO’donnell DA, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett J (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Lee JS, Shin YK, Yoon JH, Takeuchi M, Pyun YR, Park YH (2001) Sphingomonas aquatilis sp. nov., Sphingomonas koreensis sp. nov. and Sphingomonas taejonensis sp. nov., yellow-pigmented bacteria isolated from natural mineral water. Int J Syst Evol Microbiol 51:1491–1498
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Caspermeyer J (2018) MEGA software celebrates silver anniversary. Mol Biol Evol 35:1558–1560
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian Protein Metabolism. Academic press, New York, USA, pp 21–132
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–789
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinfomatics 14:60
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407
Liu B, Liu GH, Sengonca C, Schumann P, Ge CB, Wang JP, Cui WD, Lin NQ (2015) Bacillus solani sp. nov., isolated from rhizosphere soil of a potato field. Int J Syst Evol Microbiol 65:4066–4071
Zhou Y, Xu J, Xu L, Tindall BJ (2009) Falsibacillus pallidus to replace the homonym Bacillus pallidus Zhou et al 2008. Int J Syst Evol Microbiol 59:3176–3180
Tindall BJ, Rosselló-Mora R, Busse HJ, Ludwig W, Kämpfer P (2010) Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60:49–266
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky ML, Moore LH, Moore WEC, Murray RGE (1987) International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464
Liu Y, Li N, Eom MK, Schumann P, Zhang X, Cao Y, Ge Y, Xiao M, Zhao J, Cheng C, Kim SG (2017) Bacillus ciccensis sp. nov., isolated from maize (Zea mays L.) seeds. Int J Syst Evol Microbiol 67:4606–4611
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31701835), the National Key R & D Program of China (2017YFD0201100 and 2016YFD0800605), Science and Technology Innovation Team Program (STIT 2017–1-11), and the Agrobiogenome Program (A2015-3,A2017-4) of Fujian Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors agree to the submission, and we confirm that no competing interests, both financial and personal, are with this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zheng, X., Liu, G., Wang, Z. et al. Bacillus dafuensis sp. Nov., Isolated from a Forest Soil in China. Curr Microbiol 77, 2049–2055 (2020). https://doi.org/10.1007/s00284-020-02014-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02014-2