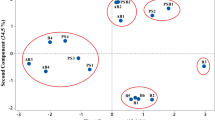
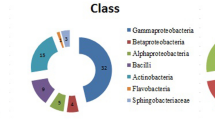
Abstract
Rhizobium are nitrogen-fixing bacteria which possess the nif gene that codes for the nitrogenase enzyme involved in the reduction of atmospheric dinitrogen (N2) to ammonia. Thirty rhizobial strains were identified from ten groundnut plant root nodules collected from semi-arid regions of Rajasthan, India. The isolates were initially identified on the basis of morphological, biochemical, and molecular characteristics. These rhizobium strains were further screened for plant growth promoting activities. Twenty-eight strains were able to produce indole acetic acid, nine strains could solubilize phosphate, and twenty-nine strains exhibited positive results for siderophore and ammonia production. All the bacterial strains were able to efficiently nodulate the groundnut under pot conditions and based on multiple PGP activities six strains were selected for field evaluation. Field experiments confirmed the effectiveness of these selected rhizobium strains resulted in significantly higher nodule number, nodule dry weight, grain yield, and yield components of inoculated plants. Inoculation of the rhizobium strain GN223 followed by GN221 resulted in high yield and field efficiency. Isolation of effective microbial strains is the prerequisite to increase the yield which is evident from the field data of the present study. Hence, these strains might serve as proficient inoculants.


Similar content being viewed by others
Abbreviations
- PCR-RFLP:
-
Polymerase chain reaction-restriction fragment length polymorphism
- ARDRA:
-
Amplified 16S ribosomal DNA restriction analysis
- UPGMA:
-
Unweighted pair group method with arithmetic mean
References
Yadav SK, Soni R, Rajput AS (2018) Role of microbes in organic farming for sustainable agro-ecosystem. In: Panpatte D, Jhala Y, Shelat H, Vyas R (eds) Microorganisms for green revolution, vol 7. Springer, Singapore, pp 241–252
Okazaki S, Nukui N, Sugawara M, Minamisawa K (2004) Rhizobial strategies to enhance symbiotic interactions: rhizobitoxine and 1-aminocyclopropane-1-carboxylate deaminase. Microbes Environ 19:99–111
Sarioglu G, Ozcelik S, Kaymaz S (1993) Selection of effective nodosity bacteria (Rhizobium leguminosarum biovar. viceae) from lentil grown in Elazıg. Turk J Agric Res 17:569–573
Rupela OP, Saxena MC (1987) Nodulation and nitrogen fixation in chickpea. In: Saxena MC, Singh KB (eds) The chickpea. CAB International, Wallingford, Oxfordshire, pp 191–206
Taurian T, Ibanez F, Fabra A, Aguilar OM (2006) Genetic diversity of rhizobia nodulating Arachis hypogaea L. in central Argentinean soils. Plant Soil 282:41–52
Yang JK, Xie FL, Zou J, Zhou Q, Zhou JC (2005) Polyphasic characteristics of Bradyrhizobia isolated from nodules of peanut (Arachis hypogaea) in China. Soil Biol Biochem 37:141–153
Sherathia D, Dey R, Thomas M, Dalsania T, Savsani K, Pal PP (2016) Biochemical and molecular characterization of DAPG-producing plant growth promoting rhizobacteria (PGPR) of groundnut (Arachis hypogaea L.). Legume Res 39(4):614–622
Li YH, Wang R, Zhang XX, Young JP, Wang ET, Sui XH, Chen WX (2015) Bradyrhizobium guangdongense sp. nov. and Bradyrhizobium guangxiense sp. nov., isolated from effective nodules of peanut. Int J Syst Evol Microbiol 65(12):4655–4661
Laguerre G, Mavingui P, Allard MR, Charnay MP, Louvrier P, Mazurier SI, Amarger N (1996) Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol 62:2029–2036
Koskey G, Mburu SW, Njeru EM, Kimiti JM, Ombori O, Maingi JM (2017) Potential of native rhizobia in enhancing nitrogen fixation and yields of climbing beans (Phaseolus vulgaris L.) in contrasting environments of eastern Kenya. Front Plant Sci 8:443
Anglade J, Billen G, Garnier J (2015) Relationships for estimating N2 fixation in legumes: incidences for N balance of legume-based cropping systems in Europe. Ecosphere 6:37
Somasegaran P, Hoben HJ (1994) Handbook for rhizobia: methods in legumes-rhizobium technology. Springer, New York, p 450
Rai R, Bantawa P, Sur S (2004) Trends in biochemical and molecular characterization of rhizobia and their nitrogen fixation mechanism: a review. In: Sen A (ed) Biology of useful plants and microbes. Narosa Publishing House, New Delhi, pp 61–119
Graham PH, Parker CA (1964) Diagnostic features in the characterization of the root-nodule bacteria of legumes. Plant Soil 20:383–396
Gordan AS, Weber RP (1951) Calorimetric estimation of indole acetic acid. Plant Physiol 26:912–915
Ali S, Charles TC, Glick BR (2014) Amelioration of high salinity stress damage by plant growth promoting bacterial endophytes that contain ACC deaminase. Plant Physiol Biochem 80:160–167
Estela M, Pachecoa H, Rodriguesa MJ, Carrilhob E, Tsaia SM (2005) Growth and siderophore production of Xylella fastidiosa under iron-limited conditions. Microbiol Res 160:429–436
Cappuccino JC, Sherman N (1992) Microbiology: a laboratory manual, 3rd edn. Benjamin/Cummings Publishing Corporation, New York, pp 125–179
Nautiyal CS (1999) An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett 170(1):265–270
Graham PH (1992) Stress tolerance in Rhizobium and Bradyrhizobium and nodulation under adverse soil conditions. Can J Microbiol 38:475–484
Hashem FM, Swelim DM, Kuykendall LD, Mohamed AI, Abdel-Wahab SM, Hegazi NI (1998) Identification and characterization of salt- and thermo-tolerant Leucaena-nodulating Rhizobium strains. Biol Fertil Soils 27:335–341
Abolhasani M, Lakzian A, Tajabadipour A, Haghnia G (2010) The study of salt and drought tolerance of Sinorhizobium bacteria to the adaptation to alkaline condition. Aust J Basic Appl Sci 4:882–886
Young JPW, Downer HL, Eardly BD (1991) Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction-based sequencing of a 16S rRNA gene segment. J Bacteriol 173:2271–2277
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol Res 173:697–703
Jain D, Sundra SD, Sanadhya S, Dhruba JN, Khandelwal SK (2017) Molecular characterization and PCR-based screening of cry genes from Bacillus thuringiensis strains. Biotechnology 7:4
Cihan CA, Tekin N, Ozcan B, Cokmus C (2012) The genetic diversity of genus Bacillus and the related genera revealed by 16s rRNA gene sequences and ARDRA analyses isolated from geothermal regions of turkey. Braz J Microbiol 43(1):309–324
Rohlf FJ (1998) NTSYSpc Numerical taxonomy and multivariate analysis system version 2.0 user guide. Applied Biostatistics Inc., Setauket, New York, p 37
Amalraj LDE, Venkateswarlu B, Desai S (2013) Effect of polymeric additives, adjuvants, surfactants on survival, stability and plant growth promoting ability of liquid bioinoculants. J Plant Physiol Pathol 1:2
Thompson JD, Higgins DG, Gibson J (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res 22(22):4673–4680
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Hewedy OA, Eissa RA, Elzanaty AM, Abd NHH, Elbary MI (2014) Phenotypic and genotypic diversity of rhizobia nodulating Faba bean from various Egyptian locations. J Bioprocess Biotechn 4:5
Agah MV, Orji JO, Nnachi AU, Chukwu OS, Udu-Ibiam OE (2016) Isolation and identification of Rhizobium species from root nodules of Arachis hypogaea L. and Telfairia occidentalis in South-east Nigeria. Int J Sci Res 5(6):227–230
Kaur J, Verma M, Lal R (2011) Rhizobium rosettiformans sp. nov., isolated from a hexachlorocyclohexane dump site, and reclassification of Blastobacter aggregatus Hirsch and Muller 1986 as Rhizobium aggregatum comb. nov. Int J Syst Evol Microbiol 61:1218–1225
Sayyed RZ, Jamadar DD, Patel RP (2011) Production of exopolysaccharide by Rhizobium sp. Indian J Microbiol 51(3):294–300
Kumari P, Meena M, Upadhyay RS (2018) Characterization of plant growth promoting rhizobacteria (PGPR) isolated from the rhizosphere of Vigna radiata (mung bean). Biocatal Agric Biotechnol 16:155–162
Sridevi M, Mallaiah KV (2009) Phosphate solubilization by Rhizobium strains. Indian J Microbiol 49(1):98–102
Karnwal A (2009) Production of indole acetic acid by fluorescent Pseudomonas in the presence of L-tryptophan and rice root exudates. J Plant Path 91:61–63
Deshwal V, Dubey R, Maheshwari D (2003) Isolation of plant growth-promoting strains of Bradyrhizobium (Arachis) sp. with biocontrol potential against Macrophomina phaseolina causing charcoal rot of peanut. Curr Sci 84:443–448
Ahmad M, Zahir ZA, Khalid M (2013) Efficacy of Rhizobium and Pseudomonas strains to improve physiology, ionic balance and quality of mung bean under salt-affected conditions on farmer’s field. Plant Physiol Biochem 63:170–176
Zahran HH, Abdel-Fattah M, Yasser MM, Mahmoud AM, Bedmar EJ (2012) Diversity and environmental stress responses of rhizobial bacteria from Egyptian grain legumes. Aust J Basic Appl Sci 6:571–583
Dong R, Zhang J, Huan H, Bai C, Chen Z, Liu G (2017) High salt tolerance of a Bradyrhizobium strain and its promotion of the growth of Stylosanthes guianensis. Int J Mol Sci 18:1625
Abdel-Lateif K, Hewedy OA, El-Zanaty AF (2016) Phylogenetic analysis of 23S rRNA gene sequences of some Rhizobium leguminosarum isolates and their tolerance to drought. Afr J Biotechnol 15:1871–1876
Koskey G, Mburu SW, Kimiti JM, Ombori O, Maingi JM, Njeru EM (2018) Genetic characterization and diversity of Rhizobium isolated from root nodules of mid-altitude climbing bean (Phaseolus vulgaris L.). Front Microbial 9:968
Santosa JWM, Silvab JFD, Ferreirac TDDS (2017) Molecular and symbiotic characterization of peanut Bradyrhizobia from the semi-arid region of Brazil. Appl Soil Ecol 121:177–184
Acknowledgements
The financial assistance from All India Network Project on soil biodiversity and biofertilizers and Rastriya Krishi Vikas Yojana (RKVY) research project are highly acknowledged. Ms. Suman Sanadhya gratefully acknowledges the CSIR for the SRF fellowship. The support of Dean, RCA and Director of Research, MPUAT is also gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
DJ conceived and designed the experiments; SS, HS, and DM performed laboratory experiments; PBS, ArS, and RHM performed field studies; DJ, AbS, RS, and SRM wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jain, D., Sanadhya, S., Saheewala, H. et al. Molecular Diversity Analysis of Plant Growth Promoting Rhizobium Isolated from Groundnut and Evaluation of Their Field Efficacy. Curr Microbiol 77, 1550–1557 (2020). https://doi.org/10.1007/s00284-020-01963-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01963-y