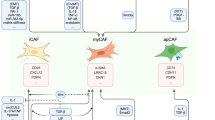
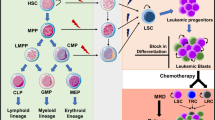
Abstract
Tumor-infiltrating T cells are promising drug targets to modulate the tumor microenvironment. However, tumor-infiltrating T lymphocytes, as central targets of cancer immunotherapy, show considerable heterogeneity and dynamics across tumor microenvironments and cancer types that may fundamentally influence cancer growth, metastasis, relapse, and response to clinical drugs. The T cell heterogeneity not only refers to the composition of subpopulations but also divergent metabolic states of T cells. Comparing to the diversity of tumor-infiltrating T cell compositions that have been well recognized, the metabolic diversity of T cells deserves more attention for precision immunotherapy. Single-cell sequencing technology enables panoramic stitching of the tumor bulk, partly by showing the metabolic-related gene expression profiles of tumor-infiltrating T cells at a single-cell resolution. Therefore, we here discuss T cell metabolism reprogramming triggered by tumor microenvironment as well as the potential application of metabolic targeting drugs. The tumor-infiltrating T cells metabolic pathway addictions among different cancer types are also addressed in this brief review.


Similar content being viewed by others
References
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100(1):57–70
Coley WB (1910) The treatment of inoperable sarcoma by bacterial toxins (the mixed toxins of the streptococcus erysipelas and the bacillus prodigiosus). Proc R Soc Med 3(Surg Sect):1–48
Peske JD, Woods AB, Engelhard VH (2015) Control of CD8 T-cell infiltration into tumors by vasculature and microenvironment. Adv Cancer Res 128:263–307
Calì B, Molon B, Viola A (2017) Tuning cancer fate: the unremitting role of host immunity. Open Biol 7(4):170006
Park R, Winnicki M, Liu E, Chu WM (2019) Immune checkpoints and cancer in the immunogenomics era. Brief Funct Genomics 18(2):133–139
Kambayashi Y, Fujimura T, Hidaka T, Aiba S (2019) Biomarkers for predicting efficacies of anti-PD1 antibodies. Front Med (Lausanne) 6:174
Sun C, Mezzadra R, Schumacher TN (2018) Regulation and function of the PD-L1 checkpoint. Immunity 48(3):434–452
Rowshanravan B, Halliday N, Sansom DM (2018) CTLA-4: a moving target in immunotherapy. Blood 131(1):58–67
Balch CM, Riley LB, Bae YJ et al (1990) Patterns of human tumor-infiltrating lymphocytes in 120 human cancers. Arch Surg 125(2):200–205
Solis-Castillo LA, Garcia-Romo GS, Diaz-Rodriguez A et al (2020) Tumor-infiltrating regulatory T cells, CD8/Treg ratio, and cancer stem cells are correlated with lymph node metastasis in patients with early breast cancer. Breast Cancer 27(5):837–849
Sharabi A, Tsokos GC (2020) T cell metabolism: new insights in systemic lupus erythematosus pathogenesis and therapy. Nat Rev Rheumatol 16(2):100–112
Kishton RJ, Sukumar M, Restifo NP (2017) Metabolic regulation of T cell longevity and function in tumor immunotherapy. Cell Metab 26(1):94–109
Alcazar I, Marques M, Kumar A et al (2007) Phosphoinositide 3-kinase gamma participates in T cell receptor-induced T cell activation. J Exp Med 204(12):2977–2987
Wang R, Dillon CP, Shi LZ et al (2011) The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35(6):871–882
Chou C, Pinto AK, Curtis JD et al (2014) c-Myc-induced transcription factor AP4 is required for host protection mediated by CD8+ T cells. Nat Immunol 15(9):884–893
Macintyre AN, Gerriets VA, Nichols AG et al (2014) The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab 20(1):61–72
Carr EL, Kelman A, Wu GS et al (2010) Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol 185(2):1037–1044
Maciver NJ, Jacobs SR, Wieman HL, Wofford JA, Coloff JL, Rathmell JC (2008) Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J Leukoc Biol 84(4):949–957
Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Rathmell JC (2018) Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175(7):1780–1795
Pacella I, Procaccini C, Focaccetti C et al (2018) Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc Natl Acad Sci U S A 115(28):E6546–E6555
Berod L, Friedrich C, Nandan A et al (2014) De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat Med 20(11):1327–1333
O’Sullivan D, van der Windt GJW, Huang SC et al (2018) Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity 49(2):375–376
Raud B, Roy DG, Divakaruni AS et al (2018) Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab 28(3):504–515
Ma R, Ji T, Zhang H et al (2018) A Pck1-directed glycogen metabolic program regulates formation and maintenance of memory CD8(+) T cells. Nat Cell Biol 20(1):21–27
van der Windt GJ, Everts B, Chang CH et al (2012) Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36(1):68–78
O’Sullivan D (2019) The metabolic spectrum of memory T cells. Immunol Cell Biol 97(7):636–646
Phan AT, Doedens AL, Palazon A et al (2016) Constitutive glycolytic metabolism supports CD8(+) T cell effector memory differentiation during viral infection. Immunity 45(5):1024–1037
Sukumar M, Liu J, Ji Y et al (2013) Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest 123(10):4479–4488
Schenkel JM, Masopust D (2014) Tissue-resident memory T cells. Immunity 41(6):886–897
Han SJ, Glatman Zaretsky A, Andrade-Oliveira V et al (2017) White adipose tissue is a reservoir for memory T cells and promotes protective memory responses to infection. Immunity 47(6):1154–1168
Chapman NM, Zeng H, Nguyen TM et al (2018) mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat Commun 9(1):2095
Pacella I, Piconese S (2019) Immunometabolic checkpoints of Treg dynamics: adaptation to microenvironmental opportunities and challenges. Front Immunol 10:1889
Jin M, Cao W, Chen B, Xiong M, Cao G (2022) Tumor-derived lactate creates a favorable niche for tumor via supplying energy source for tumor and modulating the tumor microenvironment. Front Cell Dev Biol 10:808859
Holm E, Hagmuller E, Staedt U et al (1995) Substrate balances across colonic carcinomas in humans. Cancer Res 55(6):1373–1378
San-Millan I, Brooks GA (2017) Reexamining cancer metabolism: lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 38(2):119–133
Feng Q, Liu Z, Yu X et al (2022) Lactate increases stemness of CD8 + T cells to augment anti-tumor immunity. Nat Commun 13(1):4981
Watson MJ, Vignali PDA, Mullett SJ et al (2021) Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature 591(7851):645–651
Ippolito L, Morandi A, Taddei ML et al (2019) Cancer-associated fibroblasts promote prostate cancer malignancy via metabolic rewiring and mitochondrial transfer. Oncogene 38(27):5339–5355
Xiao Z, Dai Z, Locasale JW (2019) Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun 10(1):3763
Pan M, Reid MA, Lowman XH et al (2016) Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat Cell Biol 18(10):1090–1101
Bott AJ, Maimouni S, Zong WX (2019) The pleiotropic effects of glutamine metabolism in Cancer. Cancers 11(6):770
Commisso C, Davidson SM, Soydaner-Azeloglu RG et al (2013) Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497(7451):633–637
Ron-Harel N, Ghergurovich JM, Notarangelo G et al (2019) T cell activation depends on extracellular alanine. Cell Rep 28(12):3011–3021
Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA (2013) Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol 14(5):500–508
Sharma MD, Baban B, Chandler P et al (2007) Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J Clin Invest 117(9):2570–2582
Dugnani E, Pasquale V, Bordignon C, Canu A, Piemonti L, Monti P (2017) Integrating T cell metabolism in cancer immunotherapy. Cancer Lett 411:12–18
Zhao Q, Chu Z, Zhu L et al (2017) 2-Deoxy-d-Glucose treatment decreases anti-inflammatory M2 macrophage polarization in mice with tumor and allergic airway inflammation. Front Immunol 8:637
Kagoya Y, Nakatsugawa M, Yamashita Y et al (2016) BET bromodomain inhibition enhances T cell persistence and function in adoptive immunotherapy models. J Clin Invest 126(9):3479–3494
Pearce EL, Walsh MC, Cejas PJ et al (2009) Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460(7251):103–107
Nabe S, Yamada T, Suzuki J et al (2018) Reinforce the antitumor activity of CD8(+) T cells via glutamine restriction. Cancer Sci 109(12):3737–3750
Zhong L, Li Y, Muluh TA, Wang Y (2023) Combination of CAR-T cell therapy and radiotherapy: opportunities and challenges in solid tumors (Review). Oncol Lett 26(1):281
Xu X, Gnanaprakasam JNR, Sherman J, Wang R (2019) A metabolism toolbox for CAR T therapy. Front Oncol 9:322
Vormittag P, Gunn R, Ghorashian S, Veraitch FS (2018) A guide to manufacturing CAR T cell therapies. Curr Opin Biotechnol 53:164–181
Fan TW, Warmoes MO, Sun Q et al (2016) Distinctly perturbed metabolic networks underlie differential tumor tissue damages induced by immune modulator beta-glucan in a two-case ex vivo non-small-cell lung cancer study. Cold Spring Harb Mol Case Stud 2(4):a000893
Vande Voorde J, Ackermann T, Pfetzer N et al (2019) Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci Adv 5(1):eaau7314
Funding
LJJ is supported by the National Natural Science Foundation of China (Grant No. 82104289), Science and Technology Innovation Plan from Weifang Medical University (041004), Yuandu Scholar Grant of Weifang City to LJJ, Weifang Science and Technology Bureau Plan Project (2021YX081), Shandong Province Traditional Chinese Medicine Science and Technology Project (M-2022053) and Postdoctoral Research Grant of Shenzhen to LPP (2237PT).
Author information
Authors and Affiliations
Contributions
Peipei Li, Fangchao Li and Xiaoyang Yu wrote the main manuscript text, Yanfei Zhang prepared figures and Jingjing Li designed and revised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, P., Li, F., Zhang, Y. et al. Metabolic diversity of tumor-infiltrating T cells as target for anti-immune therapeutics. Cancer Immunol Immunother 72, 3453–3460 (2023). https://doi.org/10.1007/s00262-023-03540-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-023-03540-1