Abstract
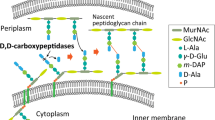
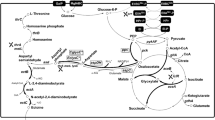
D-p-hydroxyphenylglycine (D-HPG) functions as an intermediate and has important value in antibiotic industries. The high pollution and costs from chemical processes make biotechnological route for D-HPG highly desirable. Here, a whole-cell transformation process by D-hydantoinase(Hase) and D-carbamoylase(Case) was developed to produce D-HPG from DL-hydroxyphenylhydantoin(DL-HPH) in Escherichia coli. The artificially designed ribosome binding site with strong intensity significantly facilitated the protein expression of limiting step enzyme Case. Next, the cell wall permeability was improved by disturbing the peptidoglycan structure by overproduction of D,D-carboxypeptidases without obviously affecting cell growth, to increase the bioavailability of low soluble hydantoin substrate. By fine-tuning regulation of expression level of D,D-carboxypeptidase DacB, the final production yield of D-HPG increased to 100% with 140 mM DL-HPH substrate under the optimized transformation conditions. This is the first example to enhance bio-productivity of chemicals by cell wall engineering and creates a new vision on biotransformation of sparingly soluble substrates. Additionally, the newly demonstrated ‘hydroxyl occupancy’ phenomenon when Case reacts with hydroxyl substrates provides a referential information for the enzyme engineering in future.






Similar content being viewed by others
References
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2:19–25
Andreas SB, Michael S, Karlheinz D (1998) Biocatalysis to amino acid-based chiral pharmaceuticals-examples and perspectives. J Mol Catal B-Enzym 5(1-4):1–11
Beveridge TJ (1999) Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol 181(16):4725–4733
Cai YH, Trodler P, Jiang S, Zhang W, Wu Y, Lu YH, Yang S, Jiang WH (2009) Isolation and molecular characterization of a novel D-hydantoinase from Jannaschia sp. CCS1. FEBS J 276(13):3575–3588
Cescau S, Debarbieux L, Wandersman C (2007) Probing the in vivo dynamics of Type I protein secretion complex association through sensitivity to detergents. J Bacteriol 189(5):1496–1504
Chao YP, Fu H, Lo TE, Chen PT, Wang JJ (1999)One-step production of D-p-hydroxyphenylglycine by recombinant Escherichia coli strains. Biotechnol Prog 15(6):1039–1045
Chen XY, Zaro JL, Shen WC (2013) Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev 65(10):1357–1369
Cheon YH, Kim HS, Han KH, Abendroth J, Niefind K, Schomburg D, Wang J, Kim Y (2002) Crystal structure of D-hydantoinase from Bacillus stearothermophilus: insight into the stereochemistry of enantioselectivity. Biochemistry 41(30):9410–9417
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: An N•log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Denome SA, Elf PK, Henderson TA, Nelson DE, Young KD (1999)Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J Bacteriol 181(13):3981–3993
Fan CH, Lee CK, Chao YP (2000) Recombinant Escherichia coli cell for D-p-hydroxyphenylglycine production from D-N-carbamoyl-p- hydroxyphenylglycine. Enzym Microb Technol 26(2-4):222–228
Horne D, Hakenbeck R, Tomasz A (1977) Secretion of lipids induced by inhibition of peptidoglycan synthesis in streptococci. J Bacteriol 132(2):704–717
Ikenaka Y, Nanba H, Yamada Y, Yajima K, Takano M, Takahashi S (1998) Screening, characterization, and cloning of the gene for N-carbamyl-D-amino acid amidohydrolase from thermotolerant soil bacteria. Biosci Biotechnol Biochem 62(5):882–886
Jiang S, Li CH, Zhang WW, Cai YH, Yang YL, Yang S, Jiang WH (2007) Directed evolution and structural analysis of N-carbamoyl-D-amino acid amidohydrolase provide insights into recombinant protein solubility in Escherichia coli. Biochem J 402(3):429–437
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Kim GJ, Kim HS (1995) Optimization of the enzymatic synthesis of D-p-hydroxyphenylglycine from DL-5-substituted hydantoin using D-hydantoinase and N-carbamoylase. Enzym Microb Technol 17(1):63–67
Kishida H, Unzai S, Roper DI, Lloyd A, Park SY, Tame JR (2006) Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics. Biochemistry 45(3):783–792
Lapointe G, Leblanc D, Morin A (1995) Use of a polymerase-chain-reaction-amplified DNA probe from Pseudomonas putida to detect D-hydantoinase-producing microorganisms by direct colony hybridization. Appl Microbiol Biotechnol 42(6):895–900
Lee DC, Kim HS (1998) Optimization of a heterogeneous reaction system for the production of optically active D-amino acids using thermostable D-hydantoinase. Biotechnol Bioeng 60(6):729–738
Lee SG, Lee DC, Hong SP, Sung MH, Kim HS (1995) Thermostable D-hydantoinase from thermophilic Bacillus stearothermophilus SD-1: characteristics of purified enzyme. Appl Microbiol Biotechnol 43(2):270–276
Lehrer RI, Barton A, Ganz T (1988) Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J Immunol Methods 108(1-2):153–158
Liu YQ, Li Q, Hu XJ, Yang JC (2008) Construction and co-expression of polycistronic plasmid encoding D-hydantoinase and D-carbamoylase for the production of D-amino acids. Enzym Microb Technol 42(7):589–593
Liu YF, Xu GC, Han RZ, Dong JJ, Ni Y (2017) Identification of D-carbamoylase for biocatalytic cascade synthesis of D-tryptophan featuring high enantioselectivity. Bioresour Technol 249:720–728
Liu B, Xiang SM, Zhao G, Wang BJ, Ma YH, Liu WF, Tao Y (2019) Efficient production of 3-hydroxypropionate from fatty acids feedstock in Escherichia coli. Metab Eng 51:121–130
Loh B, Grant C, Hancock RE (1984) Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother 26(4):546–551
Louwrier A, Knowles CJ (1996) The purification and characterization of a novel D-specific carbamoylase enzyme from an Agrobacterium sp. Enzym Microb Technol 19(8):562–571
Mengin-Lecreulx D, Lemaitre B (2005) Structure and metabolism of peptidoglycan and molecular requirements allowing its detection by the Drosophila innate immune system. J Endotoxin Res 11(2):105–111
Möller A, Syldatk C, Schulze M, Wagner F (1988)Stereo- and substrate-specificity of a D-hydantoinase and a D-N-carbamyl-amino acid amidohydrolase of Arthrobacter crystallopoietes AM 2. Enzym Microb Technol 10(10):618–625
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comput Chem 30(16):2785–2791
Nakai T, Hasegawa T, Yamashita E, Yamamoto M, Kumasaka T, Ueki T, Nanba H, Ikenaka Y, Takahashi S, Sato M, Tsukihara T (2000) Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure 8(7):729–739
Nanba H, Ikenaka Y, Yamada Y, Yajima K, Takano M, Takahashi S (1998) Isolation of Agrobacterium sp. strain KNK712 that produces N-carbamyl-D-amino acid amidohydrolase, cloning of the gene for this enzyme, and properties of the enzyme. Biosci Biotechnol Biochem 62(5):875–881
Nandanwar H, Prajapati R, Hoondal GS (2013) (D)-p-Hydroxyphenylglycine production by thermostable D-hydantoinase from Brevibacillus parabrevis-PHG1. Biocatal Biotransform 31(1):22–32
Nanninga N (1998) Morphogenesis of Escherichia coli. Microbiol Mol Biol Rev 62(1):110–129
Nelson DE, Young KD (2001) Contributions of PBP 5 and D,D-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J Bacteriol 183(10):3055–3064
Nose S (1984) A unified formulation of the constant temperature molecular-dynamics methods. J Chem Phys 81:511–519
Nowroozi FF, Baidoo EE, Ermakov S, Redding-Johanson AM, Batth TS, Petzold CJ, Keasling JD (2014) Metabolic pathway optimization using ribosome binding site variants and combinatorial gene assembly. Appl Microbiol Biotechnol 98(4):1567–1581
Ogawa J, Shimizu S, Yamada H (1993)N-carbamoyl-D-amino acid amidohydrolase from Comamonas sp. E222c purification and characterization. Eur J Biochem 212(3):685–691
Olivieri R, Fascetti E, Angelini L, Degen L (1979) Enzymatic conversion of N- carbamoyl-D-amino acids to D-amino acids. Enzym Microb Technol 1(3):201–204
Olivieri R, Fascetti E, Angelini L, Degen L (1981) Microbial transformation of racemic hydantoins to D-amino acids. Biotechnol Bioeng 23(10):2173–2183
Park JH, Kim GJ, Lee SG, Kim HS (1998) Biochemical properties of thermostable D-hydantoinase from Bacillus thermocatenulatus GH-2. Ann N Y Acad Sci 864(1):337–340
Park JH, Kim GJ, Kim HS (2000) Production of D-amino acid using whole cells of recombinant Escherichia coli with separately and coexpressed D-hydantoinase and N-carbamoylase. Biotechnol Prog 16(4):564–570
Parrinello M, Rahman A (1981) Polymorphic transitions in single-crystals - a new molecular-dynamics method. J Appl Physiol 52:7182–7190
Qian JQ, Chen CC, Liu M, Lv BF (2012) Bioconversion production D-p-hydrophenyglycine from DL-p-hydroxyphenylhydantoin by Pseudomonas putida in aqueous two-phase system. Indian J Biotechnol 11:445–452
Runser SM, Meyer PC (1993) Purification and biochemical characterization of the hydantoin hydrolyzing enzyme from Agrobacterium species. A hydantoinase with no 5,6-dihydropyrimidine amidohydrolase activity. Eur J Biochem 213(3):1315–1324
Schüttelkopf AW, van Aalten DM (2004) PRODRG - a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D60:1355–1363
Sorin EJ, Pande VS (2005) Exploring the Helix-Coil transition via all-atom equilibrium ensemble simulations. Biophys J 88(4):2472–2493
Typas A, Banzhaf M, Gross CA, Vollmer W (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10(2):123–136
Vollmer W, Blanot D, de Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32(2):149–167
Wu S, Liu Y, Liu YB, Zhao GG, Wang JJ, Sun WR (2005) Enzymatic production of D-p-hydroxyphenylglycine from DL-5-p-hydroxyphenylhydantoin by Sinorhizobium morelens S-5. Enzym Microb Technol 36(4):520–526
Wu H, Chen J, Chen GQ (2016) Engineering the growth pattern and cell morphology for enhanced PHB production by Escherichia coli. Appl Microbiol Biotechnol 100(23):9907–9916
Xu Z, Liu YQ, Yang YL, Jiang WH, Arnold E, Ding JP (2003) Crystal structure of D-hydantoinase from Burkholderia pickettii at a resolution of 2.7 Angstroms: insights into the molecular basis of enzyme thermostability. J Bacteriol 185(14):4038–4049
Yang HQ, Lu X, Hu JY, Chen Y, Shen W, Liu L (2018a) Boosting secretion of extracellular protein by Escherichia coli via cell wall perturbation. Appl Environ Microbiol 84(20):e01382–e01318
Yang HQ, Hu JY, Lu X, Wang FX, Shen W, Hu W, Wang LL, Chen XZ, Liu L (2018b) Improving extracellular protein production in Escherichia coli by overexpressing D,D-carboxypeptidase to perturb peptidoglycan network synthesis and structure. Appl Microbiol Biotechnol 103(2):793–806
Yu H, Yang S, Jiang W, Yang Y (2009) Efficient biocatalytic production of D-4-hydroxyphenylglycine by whole cells of recombinant Ralstonia pickettii. Folia Microbiol 54(6):509–515
Zhang DL, Zhu FY, Fan WC, Tao RS, Yu H, Yang YL, Jiang WH, Yang S (2011) Gradually accumulating beneficial mutations to improve the thermostability of N-carbamoyl-d-amino acid amidohydrolase by step-wise evolution. Appl Microbiol Biotechnol 90(4):1361–1371
Zhang SS, Zhao XJ, Tao Y, Lou CB (2015) A novel approach for metabolic pathway optimization: Oligo-linker mediated assembly (OLMA) method. J Biol Eng 9:23
Funding
The work was supported by Beijing Natural Science Foundation, China (5182021), National Science Foundation of China (11604359), and Ministry of Science & Technology, China (KY201701011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Y., Zhu, L., Qi, W. et al. Biocatalytic production of D-p-hydroxyphenylglycine by optimizing protein expression and cell wall engineering in Escherichia coli. Appl Microbiol Biotechnol 103, 8839–8851 (2019). https://doi.org/10.1007/s00253-019-10155-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10155-z