Abstract
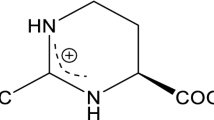
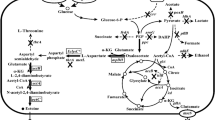
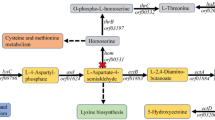
Ectoine is a high-value stabilizer and protective agent with various applications in enzyme industry, cosmetics, and biomedicine. In this study, rational engineering strategies have been implemented in Escherichia coli to efficiently produce ectoine. First, the synthetic pathway of ectoine was constructed in E. coli MG1655 by introducing an artificial thermal switch system harboring the ectABC cluster from Halomonas elongate, and the resulting strain produced 1.95 g/L ectoine. Second, crr encoding the glucose-specific enzyme II domain A of phosphotransferase system and iclR encoding the glyoxylate shunt transcriptional repressor were deleted in E. coli for enhancing the oxaloacetate supply, leading to the increasement of the ectoine titer to 9.09 g/L. Third, thrA encoding the bifunctional aspartokinase/homoserine dehydrogenase was removed from the genome to weaken the competitive pathway; simultaneously, an endogenous feedback-resistant lysC was overexpressed to complement the enzymatic activity deficiency of the aspartate kinase, leading to 30.36% increase of ectoine titer. Next, the expression of phosphoenolpyruvate carboxylase was modulated with varying gradient strength promoters to accelerate the biosynthesis efficiency of ectoine. Finally, aspDH encoding aspartate dehydrogenase from Pseudomonas aeruginosa PAO1 was overexpressed to further improve the biosynthesis of ectoine. The final strain MWZ003/pFT28-ectABC-EclysC*-aspDH-ppc3 produced 30.37 g/L ectoine after 36-h fed-batch fermentation with a yield of 0.132 g/g glucose and a productivity of 0.844 g/(L h).






Similar content being viewed by others
Availability of data and material
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request. In addition, the GeneBank accession number of the 2433-bp codon-optimized ectABC gene cluster is MW316739.
Code availability
Not applicable.
References
Galinski EA, Pfeiffer HP, Trüper HG. 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur J Biochem. 1985;149:135–9.
Lentzen G, Schwarz T. Extremolytes: natural compounds from extremophiles for versatile applications. Appl Microbiol Biotechnol. 2006;72:623–34.
Graf R, Anzali S, Buenger J, Pfluecker F, Driller H. The multifunctional role of ectoine as a natural cell protectant. Clin Dermatol. 2008;26:326–33.
Kanapathipillai M, Lentzen G, Sierks M, Park CB. Ectoine and hydroxyectoine inhibit aggregation and neurotoxicity of Alzheimer’s beta-amyloid. FEBS Lett. 2005;579:4775–80.
Sydlik U, Weissenberg A, Peuschel H, Krutmann J, Unfried K. The compatible solute ectoine protects against nanoparticle-induced neutrophilic lung inflammation. Toxicol Lett. 2009;189:S188–S188.
Abdel-Aziz H, Wadie W, Abdallah DM, Lentzen G, Khayyal MT. Novel effects of ectoine, a bacteria-derived natural tetrahydropyrimidine, in experimental colitis. Phytomedicine. 2013;20:585–91.
Czech L, Hermann L, Stöveken N, Richter AA, Höppner A, Smits SHJ, Heider J, Bremer E. Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: genetics, phylogenomics, biochemistry, and structural analysis. Genes. 2018;9:177.
Pastor JM, Salvador M, Argandona M, Bernal V, Reina-Bueno M, Csonka LN, Iborra JL, Vargas C, Nieto JJ, Canovas M. Ectoines in cell stress protection: uses and biotechnological production. Biotechnol Adv. 2010;28:782–801.
Sauer T, Galinski EA. Bacterial milking: a novel bioprocess for production of compatible solutes. Biotechnol Bioeng. 1998;57:306–13.
Fallet C, Rohe P, Franco-Lara E. Process optimization of the integrated synthesis and secretion of ectoine and hydroxyectoine under hyper/hypo-osmotic stress. Biotechnol Bioeng. 2010;107:124–33.
Schubert T, Maskow T, Benndorf D, Harms H, Breuer U. Continuous synthesis and excretion of the compatible solute ectoine by a transgenic, nonhalophilic bacterium. Appl Environ Microbiol. 2007;73:3343–7.
Grammann K, Volke A, Kunte HJ. New type of osmoregulated solute transporter identified in halophilic members of the Bacteria domain: TRAP transporter TeaABC mediates uptake of ectoine and hydroxyectoine in Halomonas elongata DSM 2581(T). J Bacteriol. 2002;184:3078–85.
Tetsch L, Kunte HJ. The substrate-binding protein TeaA of the osmoregulated ectoine transporter TeaABC from Halomonas elongata: purification and characterization of recombinant TeaA. FEMS Microbiol Lett. 2002;211:213–8.
Zhang L-H, Lang Y-J, Nagata S. Efficient production of ectoine using ectoine-excreting strain. Extremophiles. 2009;13:717–24.
Pérez-García F, Ziert C, Risse JM, Wendisch VF. Improved fermentative production of the compatible solute ectoine by Corynebacterium glutamicum from glucose and alternative carbon sources. J Biotechnol. 2017;258:59–68.
Becker J, Schäfer R, Kohlstedt M, Harder BJ, Borchert NS, Stöveken N, Bremer E, Wittmann C. Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb Cell Fact. 2013;12:110–110.
Giesselmann G, Dietrich D, Jungmann L, Kohlstedt M, Jeon E, Yim SS, Sommer F, Zimmer D, Mühlhaus T, Schroda M, Jeong K, Becker J, Wittmann C. Metabolic engineering of Corynebacterium glutamicum for high-level ectoine production: design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway. Biotechnol J. 2019;14:e201800417.
He Y-Z, Gong J, Yu H-Y, Tao Y, Zhang S, Dong Z-Y. High production of ectoine from aspartate and glycerol by use of whole-cell biocatalysis in recombinant Escherichia coli. Microb Cell Fact. 2015;14:55.
Chen W, Zhang S, Jiang P, Yao J, He Y, Chen L, Gui X, Dong Z, Tang S-Y. Design of an ectoine-responsive AraC mutant and its application in metabolic engineering of ectoine biosynthesis. Metab Eng. 2015;30:149–55.
Ning Y, Wu X, Zhang C, Xu Q, Chen N, Xie X. Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli. Metab Eng. 2016;36:10–8.
Ma H, Zhao Y, Huang W, Zhang L, Wu F, Ye J, Chen G-Q. Rational flux-tuning of Halomonas bluephagenesis for co-production of bioplastic PHB and ectoine. Nat Commun. 2020;11:3313–3313.
Plumbridge J. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr Opin Microbiol. 2002;5:187–93.
Zhu LF, Fang Y, Ding ZX, Zhang SY, Wang XY. Developing an l-threonine-producing strain from wild-type Escherichia coli by modifying the glucose uptake, glyoxylate shunt, and l-threonine biosynthetic pathway. Biotechnol Appl Biochem. 2019;15:962–76.
Zhao H, Fang Y, Wang XY, Zhao L, Wang JL, Li Y. Increasing l-threonine production in Escherichia coli by engineering the glyoxylate shunt and the l-threonine biosynthesis pathway. Appl Microbiol Biotechnol. 2018;102:5505–18.
Song CW, Lee J, Ko Y-S, Lee SY. Metabolic engineering of Escherichia coli for the production of 3-aminopropionic acid. Metab Eng. 2015;30:121–9.
Zou X, Guo L, Huang L, Li M, Zhang S, Yang A, Zhang Y, Zhu L, Zhang H, Zhang J, Feng Z. Pathway construction and metabolic engineering for fermentative production of β-alanine in Escherichia coli. Appl Microbiol Biotechnol. 2020;104:2545–59.
Lee KH, Park JH, Kim TY, Kim HU, Lee SY. Systems metabolic engineering of Escherichia coli for Kl-threonine production. Mol Syst Biol. 2007;3:149–149.
Li Y, Kawakami N, Ogola HJO, Ashida H, Ishikawa T, Shibata H, Sawa Y. A novel l-aspartate dehydrogenase from the mesophilic bacterium Pseudomonas aeruginosa PAO1: molecular characterization and application for l-aspartate production. Appl Microbiol Biotechnol. 2011;90:1953–62.
Li YJ, Wei HB, Wang T, Xu QY, Zhang CL, Fan XG, Ma Q, Chen N, Xie XX. Current status on metabolic engineering for the production of l-aspartate family amino acids and derivatives. Bioresour Technol. 2017;245:1588–602.
Wendisch VF. Metabolic engineering advances and prospects for amino acid production. Metab Eng. 2020;58:17–34.
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–14.
Fang Y, Wang J, Ma W, Yang J, Zhang H, Zhao L, Chen S, Zhang S, Hu X, Li Y, Wang X. Rebalancing microbial carbon distribution for l-threonine maximization using a thermal switch system. Metab Eng. 2020;61:33–46.
Yang J, Fang Y, Wang J, Wang C, Zhao L, Wang X. Deletion of regulator-encoding genes fadR, fabR and iclR to increase l-threonine production in Escherichia coli. Appl Microbiol Biotechnol. 2019;103:4549–64.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Zhang W, Liu Z, Gong M, Li N, Lv X, Dong X, Liu Y, Li J, Du G, Liu L. Metabolic engineering of Escherichia coli for the production of lacto-N-neotetraose (LNnT). Syst Microbiol Biomanuf. 2021. https://doi.org/10.1007/s43393-021-00023-1.
Sugino Y, Morita M. Cold-sensitive phenotype of Escherichia coli cells harboring a plasmid carrying the kil gene of phage lambda brought under control of cI857 gene promoters. Gene. 1994;141:25–30.
Becker J, Zelder O, Häfner S, Schröder H, Wittmann C. From zero to hero—design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng. 2011;13:159–68.
Rédei GP. Glucose effect. In: Rédei GP, editor. Encyclopedia of genetics, genomics, proteomics and informatics. Dordrecht: Springer; 2008. p. 803–4.
Funding
This study was supported by the National Key R&D Program of China (2018YFA0900300), the National First-class Discipline Program of Light Industry Technology and Engineering (LITE2018-10), the Key Technology Project of Inner Mongolia Autonomous Region in China (2019GG302), and the Collaborative Innovation Center of Jiangsu Modern Industrial Fermentation.
Author information
Authors and Affiliations
Contributions
SZ, YF and XW conceived and designed the research. SZ, YF, LZ, HL, ZW, and YL conducted the experiments. SZ, YF and XW analyzed the data and wrote the manuscript. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all the authors, the corresponding author states that there is no conflict of interest.
Compliance with ethics requirements
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Zhang, S., Fang, Y., Zhu, L. et al. Metabolic engineering of Escherichia coli for efficient ectoine production. Syst Microbiol and Biomanuf 1, 444–458 (2021). https://doi.org/10.1007/s43393-021-00031-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-021-00031-1