Abstract
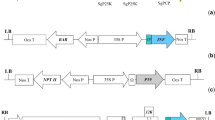
Ti and Ri plasmids of pathogenic Agrobacterium strains are stably maintained by the function of a repABC operon and have been classified into four incompatibility groups, namely, incRh1, incRh2, incRh3, and incRh4. Removal of these plasmids from their bacterial cells is an important step in determining strain-specific virulence characteristics and to construct strains useful for transformation. Here, we developed two powerful tools to improve this process. We first established a reporter system to detect the presence and absence of Ti/Ri plasmids in cells by using an acetosyringone (AS)-inducible promoter of the Ti2 small RNA and luxAB from Vibrio harveyi. This system distinguished a Ti/Ri plasmid-free cell colony among plasmid-harboring cell colonies by causing the latter colonies to emit light in response to AS. We then constructed new “Ti/Ri eviction plasmids,” each of which carries a repABC from one of four Ti/Ri plasmids that belonged to incRh1, incRh2, incRh3, and incRh4 groups in the suicidal plasmid pK18mobsacB and in a broad-host-range pBBR1 vector. Introduction of the new eviction plasmids into Agrobacterium cells harboring the corresponding Ti/Ri plasmids led to Ti/Ri plasmid-free cells in every incRh group. The Ti/Ri eviction was more effective by plasmids with the pBBR1 backbone than by those with the pK18mobsacB backbone. Furthermore, the highly stable cryptic plasmid pAtC58 in A. tumefaciens C58 was effectively evicted by the introduction of a pBBR1 vector containing the repABC of pAtC58. These results indicate that the set of pBBR1-repABC plasmids is a powerful tool for the removal of stable rhizobial plasmids.



Similar content being viewed by others
References
Austin S, Nordström K (1990) Partition-mediated incompatibility of bacterial plasmids. Cell 60(3):351–354. https://doi.org/10.1016/0092-8674(90)90584-2
Baldwin TO, Berends T, Bunch TA, Holzman TF, Rausch SK, Shamansky L, Treat ML, Ziegler MM (1984) Cloning of the luciferase structural genes from Vibrio harveyi and expression of bioluminescence in Escherichia coli. Biochemistry 23(16):3663–3667. https://doi.org/10.1021/bi00311a014
Barton BM, Harding GP, Zuccarelli AJ (1995) A general method for detecting and sizing large plasmids. Anal Biochem 226(2):235–240. https://doi.org/10.1006/abio.1995.1220
Boivin R, Chalifour FP, Dion P (1988) Construction of a Tn5 derivative encoding bioluminescence and its introduction in Pseudomonas, Agrobacterium and Rhizobium. Mol Gen Genet 213(1):50–55. https://doi.org/10.1007/BF00333397
Broothaerts W, Mitchell HJ, Weir B, Kaines S, Smith LM, Yang W, Mayer JE, Roa-Rodriguez C, Jefferson RA (2005) Gene transfer to plants by diverse species of bacteria. Nature 433(7026):629–633. https://doi.org/10.1038/nature03309
Cervantes-Rivera R, Romero-López C, Berzal-Herranz A, Cevallos MA (2010) Analysis of the mechanism of action of the antisense RNA that controls the replication of the repABC plasmid p42d. J Bacteriol 192(13):3268–3278. https://doi.org/10.1128/JB.00118-10
Cervantes-Rivera R, Pedraza-López F, Pérez-Segura G, Cevallos MA (2011) The replication origin of a repABC plasmid. BMC Microbiol 11(1):1–14. https://doi.org/10.1186/1471-2180-11-158
Cevallos MA, Cervantes-Rivera R, Gutiérrez-Ríos RM (2008) The repABC plasmid family. Plasmid 60(1):19–37. https://doi.org/10.1016/j.plasmid.2008.03.001
Chai Y, Winans SC (2005) A small antisense RNA downregulates expression of an essential replicase protein of an Agrobacterium tumefaciens Ti plasmid. Mol Microbiol 56(6):1574–1585. https://doi.org/10.1111/j.1365-2958.2005.04636.x
Chilton M-D, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW (1974) Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci U S A 71(9):3672–3676. https://doi.org/10.1073/pnas.71.9.3672
Christie PJ, Gordon JE (2014) The Agrobacterium Ti plasmids. Microbiol Spectr 2(6). https://doi.org/10.1128/microbiolspec.PLAS-0010-2013
Gérard JC, Canaday J, Szegedi E, de la Salle H, Otten L (1992) Physical map of the vitopine Ti plasmid pTiS4. Plasmid 28(2):146–156. https://doi.org/10.1016/0147-619X(92)90045-C
Gonzalez V, Santamaria RI, Bustos P, Hernandez-Gonzalez I, Medrano-Soto A, Moreno-Hagelsieb G, Janga SC, Ramirez MA, Jimenez-Jacinto V, Collado-Vides J, Davila G (2006) The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc Natl Acad Sci U S A 103(10):3834–3839. https://doi.org/10.1073/pnas.0508502103
Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S (2001) Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294(5550):2323–2328. https://doi.org/10.1126/science.1066803
Hamilton RH, Fall MZ (1971) The loss of tumor-initiating ability in Agrobacterium tumefaciens by incubation at high temperature. Experientia 27(2):229–230. https://doi.org/10.1007/BF02145913
Hattori Y, Iwata K, Suzuki K, Uraji M, Ohta N, Katoh A, Yoshida K (2001) Sequence characterization of the vir region of a nopaline type Ti plasmid, pTi-SAKURA. Genes Genet Syst 76(2):121–130. https://doi.org/10.1266/ggs.76.121
Hood EE, Helmer GL, Fraley RT, Chilton MD (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol 168(3):1291–1301. https://doi.org/10.1128/jb.168.3.1291-1301.1986
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2(4):208–218. https://doi.org/10.1007/BF01977351
Hooykaas PJ, den Dulk-Ras H, Ooms G, Schilperoort RA (1980) Interactions between octopine and nopaline plasmids in Agrobacterium tumefaciens. J Bacteriol 143(3):1295–1306
Jouanin L, Tourneur J, Tourneur C, Casse-Delbart F (1986) Restriction maps and homologies of the three plasmids of Agrobacterium rhizogenes strain A4. Plasmid 16(2):124–134. https://doi.org/10.1016/0147-619X(86)90071-5
Khan SR, Gaines J, Roop RM, Farrand SK (2008) Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74(16):5053–5062. https://doi.org/10.1128/AEM.01098-08
Kiyokawa K, Yamamoto S, Sakuma K, Tanaka K, Moriguchi K, Suzuki K (2009) Construction of disarmed Ti plasmids transferable between Escherichia coli and Agrobacterium species. Appl Environ Microbiol 75(7):1845–1851. https://doi.org/10.1128/AEM.01856-08
Kiyokawa K, Yamamoto S, Sato Y, Momota N, Tanaka K, Moriguchi K, Suzuki K (2012) Yeast transformation mediated by Agrobacterium strains harboring an Ri plasmid: comparative study between GALLS of an Ri plasmid and virE of a Ti plasmid. Genes Cells 17(7):597–610. https://doi.org/10.1111/j.1365-2443.2012.01612.x
Knauf VC, Nester EW (1982) Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8(1):45–54. https://doi.org/10.1016/0147-619X(82)90040-3
Kovacs LG, Pueppke SG (1993) The chromosomal background of Agrobacterium tumefaciens Chry5 conditions high virulence on soybean. Mol Plant-Microbe Interact 6(5):601–608. https://doi.org/10.1094/MPMI-6-601
Lacroix B, Tzfira T, Vainstein A, Citovsky V (2006) A case of promiscuity: Agrobacterium’s endless hunt for new partners. Trends Genet 22(1):29–37. https://doi.org/10.1016/j.tig.2005.10.004
Maas R, Saadi S, Maas WK (1989) Properties and incompatibility behavior of miniplasmids derived from the bireplicon plasmid pCG86. Mol Gen Genet MGG 218(2):190–198. https://doi.org/10.1007/BF00331268
Moore L, Warren G, Strobel G (1979) Involvement of a plasmid in the hairy root disease of plants caused by Agrobacterium rhizogenes. Plasmid 2(4):617–626. https://doi.org/10.1016/0147-619X(79)90059-3
Morton ER, Merritt PM, Bever JD, Fuqua C (2013) Large deletions in the pAtC58 megaplasmid of Agrobacterium tumefaciens can confer reduced carriage cost and increased expression of virulence genes. Genome Biol Evol 5:1353–1364. https://doi.org/10.1093/gbe/evt095
Nair GR, Liu Z, Binns AN (2003) Reexamining the role of the accessory plasmid pAtC58 in the virulence of Agrobacterium tumefaciens strain C58. Plant Physiol 133(3):989–999. https://doi.org/10.1104/pp.103.030262
Nester EW, Kosuge T (1981) Plasmids specifying plant hyperplasias. Annu Rev Microbiol 35(1):531–565. https://doi.org/10.1146/annurev.mi.35.100181.002531
Nishikawa M, Suzuki K, Yoshida K (1990) Structural and functional stability of IncP plasmids during stepwise transmission by trans-kingdom mating: promiscuous conjugation of Escherichia coli and Saccharomyces cerevisiae. Japanese. J Genet 65:323–334
Ono NN, Tian L (2011) The multiplicity of hairy root cultures: prolific possibilities. Plant Sci 180(3):439–446. https://doi.org/10.1016/j.plantsci.2010.11.012
Otten L, Burr T, Szegedi E (2008) Agrobacterium: a disease-causing bacterium. In: Tzfira T, Citovsky V (eds) Agrobacterium: from biology to biotechnology. Springer New York, New York, pp 1–46. https://doi.org/10.1007/978-0-387-72290-0_1
Pappas KM, Cevallos MA (2011) Plasmids of the Rhizobiaceae and their role in interbacterial and transkingdom interactions. In: Biocommunication in soil microorganisms. Springer Berlin Heidelberg, Germany, pp 295–337. https://doi.org/10.1007/978-3-642-14512-4_12
Pappas KM, Winans SC (2003) The RepA and RepB autorepressors and TraR play opposing roles in the regulation of a Ti plasmid repABC operon. Mol Microbiol 49(2):441–455. https://doi.org/10.1046/j.1365-2958.2003.03560.x
Pérez-Oseguera Á, Cevallos MA (2013) RepA and RepB exert plasmid incompatibility repressing the transcription of the repABC operon. Plasmid 70(3):362–376. https://doi.org/10.1016/j.plasmid.2013.08.001
Pinto UM, Pappas KM, Winans SC (2012) The ABCs of plasmid replication and segregation. Nat Rev Microbiol 10:755–765
Ramírez-Romero MA, Soberón N, Pérez-Oseguera A, Téllez-Sosa J, Cevallos MA (2000) Structural elements required for replication and incompatibility of the Rhizobium etli symbiotic plasmid. J Bacteriol 182(11):3117–3124. https://doi.org/10.1128/JB.182.11.3117-3124.2000
Ramírez-Romero MA, Tellez-Sosa J, Barrios H, Perez-Oseguera A, Rosas V, Cevallos MA (2001) RepA negatively autoregulates the transcription of the repABC operon of the Rhizobium etli symbiotic plasmid basic replicon. Mol Microbiol 42(1):195–204
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145(1):69–73. https://doi.org/10.1016/0378-1119(94)90324-7
Slater SC, Goodner BW, Setubal JC, Goldman BS, Wood DW, Nester EW (2008) The Agrobacterium tumefaciens C58 genome. In: Tzfira T, Citovsky V (eds) Agrobacterium: from biology to biotechnology. Springer New York, New York, pp 149–181. https://doi.org/10.1007/978-0-387-72290-0_4
Soberón N, Venkova-Canova T, Ramírez-Romero MA, Téllez-Sosa J, Cevallos MA (2004) Incompatibility and the partitioning site of the repABC basic replicon of the symbiotic plasmid from Rhizobium etli. Plasmid 51(3):203–216. https://doi.org/10.1016/j.plasmid.2004.01.005
Suzuki K, Hattori Y, Uraji M, Ohta N, Iwata K, Murata K, Kato A, Yoshida K (2000) Complete nucleotide sequence of a plant tumor-inducing Ti plasmid. Gene 242(1-2):331–336. https://doi.org/10.1016/S0378-1119(99)00502-8
Suzuki K, Tanaka K, Yamamoto S, Kiyokawa K, Moriguchi K, Yoshida K (2009) Ti and Ri plasmids. In: Schwartz E (ed) Microbial megaplasmids. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 133–147. https://doi.org/10.1007/978-3-540-85467-8_6
Szegedi E, Czakó M, Otten L, Koncz CS (1988) Opines in crown gall tumours induced by biotype 3 isolates of Agrobacterium tumefaciens. Physiol Mol Plant Pathol 32(2):237–247. https://doi.org/10.1016/S0885-5765(88)80020-1
Szegedi E, Czako M, Otten L (1996) Further evidence that the vitopine-type pTi’s of Agrobacterium vitis represent a novel group of Ti plasmids. Mol Plant-Microbe Interact 9(2):139–143. https://doi.org/10.1094/MPMI-9-0139
Szittner R, Meighen E (1990) Nucleotide sequence, expression, and properties of luciferase coded by lux genes from a terrestrial bacterium. J Biol Chem 265(27):16581–16587
Tanaka K, Arafat HH, Urbanczyk H, Yamamoto S, Moriguchi K, Sawada H, Suzuki K (2009) Ability of Agrobacterium tumefaciens and A. rhizogenes strains, inability of A. vitis and A. rubi strains to adapt to salt-insufficient environment, and taxonomic significance of a simple salt requirement test in the pathogenic Agrobacterium species. J Gen Appl Microbiol 55(1):35–41. https://doi.org/10.2323/jgam.55.35
Tzfira T, Citovsky V (2006) Agrobacterium-mediated genetic transformation of plants: biology and biotechnology. Curr Opin Biotechnol 17(2):147–154. https://doi.org/10.1016/j.copbio.2006.01.009
Uraji M, Suzuki K, Yoshida K (2002) A novel plasmid curing method using incompatibility of plant pathogenic Ti plasmids in Agrobacterium tumefaciens. Genes Genet Syst 77(1):1–9. https://doi.org/10.1266/ggs.77.1
Van Larebeke N, Engler G, Holsters M, Van den Elsacker S, Zaenen I, Schilperoort RA, Schell J (1974) Large plasmid in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature 252(5479):169–170. https://doi.org/10.1038/252169a0
Venkova-Canova T, Soberón NE, Ramírez-Romero MA, Cevallos MA (2004) Two discrete elements are required for the replication of a repABC plasmid: an antisense RNA and a stem-loop structure. Mol Microbiol 54(5):1431–1444. https://doi.org/10.1111/j.1365-2958.2004.04366.x
Wendt T, Doohan F, Winckelmann D, Mullins E (2011) Gene transfer into Solanum tuberosum via Rhizobium spp. Transgenic Res 20(2):377–386. https://doi.org/10.1007/s11248-010-9423-4
White FF, Nester EW (1980) Relationship of plasmids responsible for hairy root and crown gall tumorigenicity. J Bacteriol 144(2):710–720
Wilms I, Overloper A, Nowrousian M, Sharma CM, Narberhaus F (2012) Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol 9(4):446–457. https://doi.org/10.4161/rna.17212
Wise AA, Liu Z, Binns AN (2006) Culture and maintenance of Agrobacterium strains. In: Wang K (ed) Agrobacterium protocols. Humana Press, Totowa, pp 3–14. https://doi.org/10.1385/1-59745-130-4:3
Yamamoto S, Suzuki K (2012) Development of a reinforced Ti-eviction plasmid useful for construction of Ti plasmid-free Agrobacterium strains. J Microbiol Methods 89(1):53–56. https://doi.org/10.1016/j.mimet.2012.01.019
Yamamoto S, Uraji M, Tanaka K, Moriguchi K, Suzuki K (2007) Identification of pTi-SAKURA DNA region conferring enhancement of plasmid incompatibility and stability. Genes Genet Syst 82(3):197–206. https://doi.org/10.1266/ggs.82.197
Yamamoto S, Kiyokawa K, Tanaka K, Moriguchi K, Suzuki K (2009) Novel toxin-antitoxin system composed of serine protease and AAA-ATPase homologues determines the high level of stability and incompatibility of the tumor-inducing plasmid pTiC58. J Bacteriol 191(14):4656–4666. https://doi.org/10.1128/JB.00124-09
Yamamoto S, Agustina V, Sakai A, Moriguchi K, Suzuki K (2017) An extra repABC locus in the incRh2 Ti plasmid pTiBo542 exerts incompatibility toward an incRh1 plasmid. Plasmid 90:20–29. https://doi.org/10.1016/j.plasmid.2017.02.003
Young JPW, Crossman LC, Johnston AWB, Thomson NR, Ghazoui ZF, Hull KH, Wexler M, Curson ARJ, Todd JD, Poole PS, Mauchline TH, East AK, Quail MA, Churcher C, Arrowsmith C, Cherevach I, Chillingworth T, Clarke K, Cronin A, Davis P, Fraser A, Hance Z, Hauser H, Jagels K, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, Whitehead S, Parkhill J (2006) The genome of Rhizobium leguminosarum has recognizable core and accessory components. Genome Biol 7(4):R34. https://doi.org/10.1186/gb-2006-7-4-r34
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 25850003, 24580115, and 15H04479. We thank Dr. E. Szegedi for providing bacterial strains and Dr. T. Oyama (Kyoto University) for the luxAB plasmid. Pathogenic strains isolated outside of Japan were imported with permissions issued by the Japanese Plant Quarantine Office and were handled under directions given by the same organization.
Funding
This study was funded by the JSPS (25850003, 24580115, and 15H04479).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Yamamoto, S., Sakai, A., Agustina, V. et al. Effective removal of a range of Ti/Ri plasmids using a pBBR1-type vector having a repABC operon and a lux reporter system. Appl Microbiol Biotechnol 102, 1823–1836 (2018). https://doi.org/10.1007/s00253-017-8721-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8721-7