Abstract
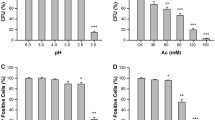
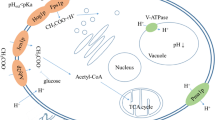
Acetic acid inhibits the metabolic activities of Saccharomyces cerevisiae. Therefore, a better understanding of how S. cerevisiae cells acquire the tolerance to acetic acid is of importance to develop robust yeast strains to be used in industry. To do this, we examined the transcriptional changes that occur at 12 h post-exposure to acetic acid, revealing that 56 and 58 genes were upregulated and downregulated, respectively. Functional categorization of them revealed that 22 protein synthesis genes and 14 stress response genes constituted the largest portion of the upregulated and downregulated genes, respectively. To evaluate the association of the regulated genes with acetic acid tolerance, 3 upregulated genes (DBP2, ASC1, and GND1) were selected among 34 non-protein synthesis genes, and 54 viable mutants individually deleted for the downregulated genes were retrieved from the non-essential haploid deletion library. Strains overexpressing ASC1 and GND1 displayed enhanced tolerance to acetic acid, whereas a strain overexpressing DBP2 was sensitive. Fifty of 54 deletion mutants displayed enhanced acetic acid tolerance. Three chosen deletion mutants (hsps82Δ, ato2Δ, and ssa3Δ) were also tolerant to benzoic acid but not propionic and sorbic acids. Moreover, all those five (two overexpressing and three deleted) strains were more efficient in proton efflux and lower in membrane permeability and internal hydrogen peroxide content than controls. Individually or in combination, those physiological changes are likely to contribute at least in part to enhanced acetic acid tolerance. Overall, information of our transcriptional profile was very useful to identify molecular factors associated with acetic acid tolerance.





Similar content being viewed by others
References
Abbott DA, Knijnenburg TA, de Poorter LM, Reinders MJ, Pronk JT, Van Maris AJ (2007) Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res 7:819–833
Almeida B, Ohlmeier S, Almeida AJ, Madeo F, Leao C, Rodrigues F, Ludovico P (2009) Yeast protein expression profile during acetic acid-induced apoptosis indicates causal involvement of the TOR pathway. Proteomics 9:720–732
Bajwa PK, Ho CY, Chan CK, Martin VJ, Trevors JT, Lee H (2013) Transcriptional profiling of Saccharomyces cerevisiae T2 cells upon exposure to hardwood spent sulphite liquor: comparison to acetic acid, furfural and hydroxymethylfurfural. Antonie Van Leeuwenhoek 103:1281–1295
Beilharz TH, Preiss T (2004) Translational profiling: the genome-wide measure of the nascent proteome. Brief Funct Genomic Proteomic 3:103–111
Carmelo V, Bogaerts P, Sá-Correia I (1996) Activity of plasma membrane H+-ATPase and expression of PMA1 and PMA2 genes in Saccharomyces cerevisiae cells grown at optimal and low pH. Arch Microbiol 166:315–320
Carmelo V, Santos H, Sá-Correia I (1997) Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim Biophys Acta 1325:63–70
Causton HC, Ren B, Koh S, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12:323–337
Fernandes AR, Mira NP, Vargas RC, Canelhas I, Sá-Correia I (2005) Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Biophys Res Commun 337:95–103
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80
Halbeisen RE, Galgano A, Scherrer T, Gerber AP (2008) Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci 65:798–813
Holyoak CD, Stratford M, McMullin Z, Cole MB, Crimmins K, Brown AJ (1996) Activity of the plasma membrane H+-ATPase and optimal glycolytic flux are required for rapid adaptation and growth of Saccharomyces cerevisiae in the presence of the weak-acid preservative sorbic acid. Appl Environ Microbiol 62:3158–3164
Kawahata M, Masaki K, Fujii T, Iefuji H (2006) Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res 6:924–936
Kim NR, Yang J, Kwon H, An J, Choi W, Kim W (2013) Mutations of the TATA-binding protein confer enhanced tolerance to hyperosmotic stress in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:3227–3228
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li BZ, Yuan YJ (2010) Transcriptome shifts in response to furfural and acetic acid in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 86:1915–1924
Marinho HS, Real C, Cyrne L, Soares H, Antunes F (2014) Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol 2:535–562
Mira NP, Becker JD, Sá-Correia I (2010a) Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS 5:587–601
Mira NP, Palma M, Guerreiro JF, Sá-Correia I (2010b) Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9:79
Mira NP, Teixeira MC, Sá-Correia I (2010c) Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 14:525–540
Mollapour M, Piper PW (2007) Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol 27:6446–6456
Mollapour M, Shepherd A, Piper PW (2008) Novel stress responses facilitate Saccharomyces cerevisiae growth in the presence of the monocarboxylate preservatives. Yeast 25:169–177
Morgan M, Anders S, Lawrence M, Aboyoun P, Pages H, Gentleman R (2009) ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25:2607–2608
Piper PW (1999) Yeast superoxide dismutase mutants reveal a prooxidant action of weak organic acid food preservatives. Free Radic Biol Med 27:1219–1227
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA
Sikkema J, de Bont JA, Poolman B (1995) Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev 59:201–222
Silva A, Sampaio-Marques B, Fernandes A, Carreto L, Rodrigues F, Holcik M, Santos MA, Ludovico P (2013) Involvement of yeast HSP90 isoforms in response to stress and cell death induced by acetic acid. PLoS One 8, e71294
Simões T, Mira NP, Fernandes AR, Sá-Correia I (2006) The SPI1 gene, encoding a glycosylphosphatidylinositol (GPI)-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak acids food preservatives. Appl Environ Microbiol 72:7168–7175
Simpson CE, Ashe MP (2012) Adaptation to stress in yeast: to translate or not? Biochem Soc Trans 40:794–799
Stevens S, Hofmeyr J-HS (1993) Effects of ethanol, octanoic and decanoic acids on fermentation and the passive influx of protons through the plasma membrane of Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38:656–663
Stratford M, Nebe-von-Caron G, Steels H, Novodvorska M, Ueckert J, Archer DB (2013) Weak-acid preservatives: pH and proton movements in the yeast Saccharomyces cerevisiae. Int J Food Microbiol 161:164–171
Szklarczyk D, FranceschiniA KM, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39:D561–D568
Tanaka K, Ishii Y, Ogawa J, Shima J (2012) Enhancement of acetic acid tolerance in Saccharomyces cerevisiae by overexpression of the HAA1 gene, encoding a transcriptional activator. Appl Environ Microbiol 78:8161–8163
Teixeira MC, Santos PM, Fernandes AR, Sá-Correia I (2005) A proteome analysis of the yeast response to the herbicide 2,4-dichlorophenoxyacetic acid. Proteomics 5:1889–1901
Yang J, Bae JY, Lee YM, Kwon H, Moon H, Kang HA, Yee S, Kim W, Choi W (2011) Construction of Saccharomyces cerevisiae strains with enhanced ethanol tolerance by mutagenesis of the TATA-binding protein gene and identification of novel genes associated with ethanol tolerance. Biotechnol Bioeng 108:1776–1787
Zheng DQ, Wu XC, Wang PM, Chi XQ, Tao XL, Li P (2011) Drug resistance marker-aided genome shuffling to improve acetic acid tolerance in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 38:415–422
Acknowledgments
We thank Eunjung Kim for support with plasmid and strain construction and Young Lee for critical reading of the manuscript. This study was partly supported by the BK21 Plus Program (Creative Academy of Ecoscience, 31Z20130012990) funded by the Ministry of Education and National Research Foundation of Korea.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 236 kb)
Rights and permissions
About this article
Cite this article
Lee, Y., Nasution, O., Choi, E. et al. Transcriptome analysis of acetic-acid-treated yeast cells identifies a large set of genes whose overexpression or deletion enhances acetic acid tolerance. Appl Microbiol Biotechnol 99, 6391–6403 (2015). https://doi.org/10.1007/s00253-015-6706-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6706-y