Abstract
Aims/hypothesis
Diabetic retinopathy is the most common complication of diabetes and a leading cause of blindness among working-age adults. Anatomical and functional changes occur in the retina and retinal pigment epithelium (RPE) prior to clinical symptoms of the disease. However, the molecular mechanisms responsible for these early changes, particularly in the RPE, remain unclear. To begin defining the molecular changes associated with pre-retinopathic diabetes, we conducted a comparative proteomics study of human donor RPE.
Methods
The RPE was dissected from diabetic human donor eyes with no clinically apparent diabetic retinopathy (n = 6) and from eyes of age-matched control donors (n = 17). Soluble proteins were separated based upon their mass and charge using two-dimensional (2-D) gel electrophoresis. Protein spots were visualised with a fluorescent dye and spot densities were compared between diabetic and control gels. Proteins from spots with significant disease-related changes in density were identified using mass spectrometry.
Results
Analysis of 325 spots on 2-D gels identified 31 spots that were either up- or downregulated relative to those from age-matched control donors. The protein identity of 18 spots was determined by mass spectrometry. A majority of altered proteins belonged to two major functional groups, metabolism and chaperones, while other affected categories included protein degradation, synthesis and transport, oxidoreductases, cytoskeletal structure and retinoid metabolism.
Conclusions/interpretation
Changes identified in the RPE proteome of pre-retinopathic diabetic donor eyes compared with age-matched controls suggest specific cellular alterations that may contribute to diabetic retinopathy. Defining the pre-retinopathic changes affecting the RPE could provide important insight into the molecular events that lead to this disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Diabetes mellitus is a multifactorial metabolic disorder that currently affects over 200 million people worldwide [1]. Models estimate that this number will nearly double by the year 2025 [1]. Diabetic retinopathy is the most common complication in diabetes and the leading cause of blindness among working-age adults [2]. The clinical manifestations of diabetic retinopathy, including retinal microaneurysms, neovascularisation, haemorrhages and macular oedema, eventually lead to visual impairment [3]. Although vascular abnormalities in the retina clearly contribute to vision loss, other anatomical and functional changes are apparent soon after the onset of diabetes [4–6]. Anatomical changes include reduced retinal thickness [4, 6] and morphological changes of several retinal cell types, including the retinal pigment epithelium (RPE) [5, 7]. Functional changes in retinal electrophysiology and contrast sensitivity have been observed in diabetic patients and animal models [6, 8, 9]. These anatomical and functional changes are apparent in the pre-retinopathic stage that precedes clinically evident vascular changes associated with the disease. Importantly, the onset of retinopathy occurs after a prolonged interval following disease onset. Klein and colleagues reported that retinopathy occurs in 73% of type 1 diabetic patients approximately 14 years after the initial diagnosis [2]. Thus, the pre-retinopathic stage provides a window of opportunity for intervention that could delay or prevent the onset of blindness. To fully maximise this potential, a thorough understanding of the biochemical changes occurring in retinal tissue is required.
The retina is composed of two major components, the neural retina and the RPE. Previous investigations have focused on the neural retina where vascular changes occur. The neural retina is composed of seven different cell types, including the photoreceptors that are responsible for visual signal transduction. The RPE is essential for vision because of its role in maintaining the photoreceptors [10]. Key functions of the RPE include regenerating the rhodopsin chromophore, regulating nutrient transport to the photoreceptors, phagocytosis of spent tips of photoreceptor outer segments [10] and the production of cytokines, including neurotrophic and angiogenic factors [11]. Additionally, the RPE provides 60 to 80% of retinal glucose via its high-capacity transport system, making it a key source of the high glucose needs of the retina [12].
To characterise early changes leading to diabetic retinopathy, several recent studies have examined changes in the proteome of the neural retina in diabetic animal models [13, 14]. The present study has focused on the RPE because of its role in supporting the retina before and after the onset of diabetes. We applied a proteomic approach using two-dimensional (2-D) gel electrophoresis and matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry to identify differences in cellular protein content in human donor RPE at the pre-retinopathic stage of diabetes.
Methods
Human tissue procurement
Donor eyes obtained from the Minnesota Lions Eye Bank (Minneapolis, MN, USA) were acquired with the consent of the donor or the donor’s family for use in medical research in accordance with the principles outlined in the Declaration of Helsinki and as approved by the University of Minnesota Institutional Review Board. Information provided by the Lions Eye Bank included sex, age, time and cause of death, and a family report of a limited medical history including ocular history and the occurrence of diabetes (Table 1).
Eyes were maintained in a moist chamber at 4°C until processing for evaluation of ocular pathology as previously outlined [15–17]. Anterior segments were removed for direct visualisation and stereoscopic fundus photography of the posterior segment. The methodology employed for imaging the posterior pole was that used in the Minnesota Grading System, which has been previously described for studies of age-related macular degeneration (AMD) [18]. The fundus, including the macula and optic nerve, was carefully examined for signs of proliferative diabetic retinopathy, microaneurysms, characteristic dot blot haemorrhages or signs of hard exudation. Some post-mortem haemorrhages are common; therefore, it is difficult to completely distinguish small intraretinal haemorrhages of early diabetic retinopathy from post-mortem changes. After reviewing eyebank eyes for over 7 years, we feel confident of being able to distinguish such haemorrhages. Post mortem haemorrhages are typically large, asymmetric, intra-, sub- or pre-retinal and regional (e.g. in one quadrant). Diabetic haemorrhages are usually small, diffuse, symmetric and specifically intraretinal. Digitised images were taken before and after removal of the neural retina for a simultaneous examination of the RPE cell layer by two ophthalmologists (T. W. Olsen and X. Feng). Eyes were excluded from the study if there was clinical evidence of retinal pathology. Eyes were also excluded if post-mortem retinal vascular changes suggested that they could be related to diabetes or the eyes had evidence of AMD. Donors with a medical history of diabetes but no signs of diabetic retinopathy were included in the pre-retinopathic group (Table 1). Three diabetic donors had type 1 and two had type 2 diabetes. The type of diabetes for one donor was not known. The age-matched control group included donors with no history of diabetes and no evidence of retinal pathology.
Protein isolation
Total RPE was dissected after collecting fundus images, pelleted by centrifugation for 30 min at 1,100 g and frozen at −80°C until processing [15]. RPE cells were fractioned by two freeze–thaw cycles and homogenised by six passes through a 26 gauge needle in a buffer containing 20 mmol/l HEPES, 10 mmol/l KCl, 1.5 mmol/l MgCl2, 250 mmol/l sucrose, 1 mmol/l EDTA, 1 mmol/l EGTA, 1 mmol/l phenylmethylsulphonyl fluoride and 0.5% NP40. Nuclei and intact cells were pelleted by centrifugation for 15 min at 600 g and 4°C, and the supernatant retained. After repeating homogenisation and centrifugation of the intact cell pellet, the first and second supernatants were combined. The supernatant from a final 15 min centrifugation at 13,000 g was stored at −80°C until use. Protein concentrations were determined using the bicinchoninic acid protein assay (Pierce Biotechnology, Rockford, IL, USA), with bovine serum albumin as the protein standard. Average protein yield for the control and diabetic eyes was 486.2 ± 34.2 μg and 558.1 ± 64.4 μg (mean ± SE), respectively. The yields were not significantly different between the two groups (p = 0.314).
2-D SDS-PAGE
The proteins extracted from the RPE of each donor were subjected to 2-D SDS PAGE (125 μg). The first dimension separation was performed with pH 5 to 8 immobilised linear gradient strips (Bio-Rad, Hercules, CA, USA). Protein samples (125 μg) were dissolved in a rehydration solution (9 mol/l urea, 3 mol/l thiourea, 6% CHAPS, 1% ASB-14, 1% Biolytes pH 3–10 (Bio-Rad) and 50 mmol/l dithiothreitol and loaded onto 11 cm IPG strips. The conditions for strip rehydration, focusing, equilibration and second-dimension separation were as outlined [17].
The 2-D gels, analysed for protein content, were stained with Flamingo fluorescent stain (Bio-Rad) according to the manufacturer’s protocol. Gels for mass spectrometry were stained with silver using a mass spectrometry-compatible kit (Silver Stain Plus Kit; Bio-Rad) and imaged with a GS-800 calibrated densitometer (Bio-Rad).
Two-dimensional gel quantification and analysis
Flamingo-stained gels were imaged at two different exposure times using a ChemiDoc XRS (Bio-Rad) and a Dark Reader Transilluminator (Clare Chemical Research, Dolores, CO, USA) to optimise the number of spots analysed. The Dark Reader was used because the light source wavelength (400–500 nm) closely matched the excitation maximum (512 nm) of the dye. Exposure times were based on the fluorescence intensity of two standard proteins run on each gel. Protein spot identification, alignment and quantification of intensity were performed using a 2-D gel analysis software (PDQuest 7.1.1; Bio-Rad). One gel with well-resolved protein spots was chosen as the master gel. Background, streaks and other staining artefacts were subtracted. Automatic spot detection and matching were followed by manual inspection and editing. Spot intensities of each gel were normalised to the total intensity of valid spots of that gel.
In-gel digestion and MALDI-TOF analysis
Spots were manually excised from 2-D silver-stained polyacrylamide gels and in-gel digestion was performed with trypsin as described [17]. Peptides were analysed by MALDI-TOF using a mass spectrometer (QSTAR XL quadrupole-TOF; Applied Biosystems, Foster City, CA, USA) located at the University of Minnesota Mass Spectrometry Consortium for the Life Sciences. All mass spectra were externally calibrated with human angiotensin II tryptic peptides (monoisotopic [MH+] 1,046.5417) and adrenocorticotropin hormone fragment (monoisotopic [MH+] 2,465.1989). Mass values corresponding to known contaminants (e.g. keratin) and to published trypsin autolysis fragments and matrix clusters were removed [19]. Monoisotopic peaks were automatically identified (Bioanalyst; Applied Biosystems) and verified by manual inspection. Peak lists were submitted to the Mascot search engine (http://www.matrixscience.com, accessed 4 March 2008) and searched in the human Swiss-Prot database (http://ca.expasy.org/sprot/, accessed 4 March 2008). Enzyme specificity was set to trypsin with no missed cleavages, a mass tolerance window of 100 ppm, carbamidomethyl as a fixed cysteine modification and oxidation of methionine as a variable modification. Initial identifications were accepted for spectra with a significant molecular weight search (MOWSE) score. Initial identities were confirmed by peptide sequencing with tandem mass spectrometry (MS/MS) using a QSTAR XL mass spectrometer (Applied Biosystems). The MS/MS spectra were submitted to Mascot set to the human Swiss-Prot database with a fragment tolerance window of 0.8 Da, peptide tolerance of 1.2 Da and MALDI-QUAD-TOF selected as the instrument. Only MS/MS spectra with a significant Mascot score (p < 0.05) were considered acceptable. The proteins were considered positively identified when the criteria of a significant full-scan match and one or more significantly matched peptide sequences were met. In some instances, a significant score derived from several peptide sequences was considered as positive verification (Tables 2 and 3).
Statistical analysis
To determine the number of samples necessary to detect statistically significant changes between groups, a power analysis was performed on Flamingo-stained gels (n = 17 control and n = 6 diabetic) as previously outlined [15]. Linear regression analysis was performed to compare spot density with time from death to freezing. Analysis was done for both gel exposure times. The p value for linear regression and critical values for correlation coefficient (R) were taken into consideration when verifying the significance of the relationship using Origin Lab 7.5 statistical software (http://www.originlab.com, accessed 4 March 2008).
The normalised intensity values of individual protein spots were compared between the two groups by Student’s two-tailed t test for unpaired samples. Values outside three interquartile ranges from the 25th and 75th percentiles of the data distribution were removed. When the assumption of equal variance (Modified Levene Equal-Variance Test) was violated, the Aspin–Welch Unequal Variance Test was used. When normality assumptions were not met, data were either transformed to natural log to obtain a normal distribution or Kolgomarov–Smirnov nonparametric test was used. The results are expressed as mean ± SEM and a value of p < 0.05 was considered statistically significant. All statistics were calculated using NCSS statistical software (NCSS 2001; Kaysville, UT, USA).
Results
Experimental design
Demographic and clinical donor information obtained from the Minnesota Lions Eye Bank is summarised in Table 1. Donors with clinically evident eye disease were excluded from the study. Average time from enucleation to freezing (17.7 ± 3.5 h; mean ± SD) was not significantly different between the two groups (p = 0.996).
Analysis included 17 and 6 gels from control and diabetic donors, respectively. A total of 325 spots were analysed. In some instances, spots were eliminated from analysis if they were not clearly resolved, had artefacts that interfered with density measurements, or were defined statistically as outliers. However, to be included in the analysis, each spot had to be present in a minimum of five samples from the diabetic group and 14 samples from the control group. A power analysis based on the average variation in intensity of individual spots from 2-D gels indicated that this number was sufficient to detect at least a 75% difference in intensity with 80% power and α = 0.05.
Linear regression was performed (spot density versus time from death to freezing) to evaluate post mortem stability of proteins in our samples. No time-dependent change in density was noted for 94% of all spots examined and no change was observed for the spots identified in our study. These results confirm that the changes in protein content in this study were not due to post mortem protein degradation, but resulted from altered protein content due to the disease state.
Analysis of protein content and protein identification
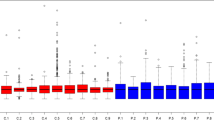
The RPE proteome was analysed using 2-D gel electrophoresis and mass spectrometry to identify differences in cellular protein content between the diabetic and control groups. A representative Flamingo-stained fluorescent gel is shown in Fig. 1. A total of 325 spots were analysed. There was a significant change in the cellular content of 31 spots with diabetes. Protein identity was determined for 18 spots. As shown in Fig. 2, 15 spots were upregulated and three were downregulated. Initial identification of protein spots was done by MALDI-TOF mass spectrometry peptide mass fingerprinting and confirmed by MS/MS peptide sequencing. Protein identification was based on six or more matching peptides (average 11, range 6–21), statistically significant MOWSE score (p < 0.05) and the sequencing of at least one peptide. Since multiple proteins can co-migrate in a single spot, we re-examined our MALDI full scans by removing the peptides that matched the identified protein and re-searching for peptides from other proteins. This secondary search did not reveal additional proteins.
RPE proteins resolved by 2-D gel electrophoresis. Representative Flamingo-stained gel (125 μg) indicates identified proteins showing altered content with diabetes. Boxed spots, increased levels; circled spots, decreased levels. The pI range for separation in the first dimension (pH 5–8) is shown at the top. The molecular mass standard is labelled in kDa on the left. Numbers correspond to density summaries in Fig. 2 and identified proteins listed in Tables 2 and 3
Summary of protein spot density. Results of densitometry for protein spots that were identified by mass spectrometry and demonstrated a significant increase (a, b) or decrease (c) in spot density. Spot numbers correspond to those in Fig. 1 and identified proteins listed in Tables 2 and 3. Dark grey bars, control; light grey bars, diabetic
Overall, the experimental molecular mass and isoelectric point (pI) were similar to the theoretical values for each protein, with the exception of cathepsin D (Table 2). Spot 6, identified as cathepsin D, migrates at an apparent mass of 31 kDa, but matched to a theoretical molecular mass of 45 kDa. This protein is initially synthesised as an inactive proenzyme (52 kDa) that is subsequently converted into an active intermediate (46 kDa) and finally cleaved in the lysosome to generate the mature 31 kDa form [20]. The migration of spot 6 is consistent with the theoretical migration of mature cathepsin D protein.
Two proteins that we identified migrated in multiple spots. Aldehyde dehydrogenase was identified in two spots (spots 2 and 3) that migrated at a similar molecular mass but a different pI, as did protein disulphide-isomerase A3 (spots 12 and 13). These findings could reflect post-translational modifications that cause an acidic or basic shift in migration for a subset of the protein population.
Classification of the identified proteins
The identified proteins were categorised according to their function on the basis of published literature and the Swiss-Prot database. The major functional groups include energy metabolism (29%) and chaperones (23%). Other functional groups include membrane dynamics (12%), structural proteins (6%), protein transport (6%), protein degradation (6%), protein synthesis (6%), retinoid metabolism (6%) and oxidoreductases (6%) (Fig. 3).
Our list includes proteins residing in different subcellular compartments. The majority of proteins were cytoplasmic (59%). Other proteins were identified from the mitochondria (aldehyde dehydrogenase, elongation factor Tu, GRP75 and succinyl CoA: 3-ketoacid-coenzyme A transferase 1), the endoplasmic reticulum (protein disulphide isomerase A3), peroxisomes (sterol carrier protein x) and lysosomes (cathepsin D).
Discussion
In the present study we analysed the RPE proteome in pre-retinopathic diabetic human donor eyes. Analysis of 325 spots on 2-D gels identified 31 spots that were either up- or downregulated relative to those from age-matched control donors. The protein identity of eighteen spots was determined by mass spectrometry. A majority of altered proteins belonged to two major functional groups, metabolism and chaperones, while other affected categories included protein degradation, synthesis and transport, oxidoreductases, cytoskeletal structure and retinoid metabolism (Fig. 3). The cellular content of approximately 62% of these proteins has previously been reported to change in response to diabetes but in non-retinal tissues (Tables 2 and 3), and thus these results represent novel findings for the retina. Because of the unique role of the RPE in supporting the photoreceptors and the retinal microenvironment, the pre-clinical changes identified here provide our first insights into the early molecular changes that precede diabetic retinopathy.
Donors of our diabetic samples had type 1 or type 2 diabetes as specified by Eye Bank records rather than by direct clinical records. Ideally, we would have analysed type 1 and 2 samples separately. However, this was not feasible because of the limited availability of tissue. If the proteomes of the two groups were significantly different in the pre-retinopathic stage, we would have observed a high variability within our diabetic group. However, no outliers were detected in this group, suggesting that the proteomes of our type 1 and type 2 diabetic donors were comparable at the pre-retinopathic stage.
The 16 proteins identified herein reflect a conservative estimate, due to our limited sample size and the technical constraints of 2-D gels. With our sample size, we were able to detect a 75% difference on average, but smaller, physiologically meaningful changes may have been undetected. Membrane proteins resolve poorly in the first dimension of a 2-D gel and are likely to be under-represented in the analysis. Low-abundance proteins are often below the detection limit of 2-D gels and would be under-represented in this analysis. While the pH range of 5 to 8 was used because it resolved the greatest number of spots, proteins outside that range would not have been detected using this methodology. Finally, due to the restricted number of tryptic peptides generated, we were unable to identify low-mass spots. Despite these caveats, this study identified many novel diabetes-related changes in the RPE proteome.
Vascular endothelial growth factor (VEGF) and pigment epithelium-derived factor (PEDF), growth factors produced by RPE, are considerably altered in diabetic retinopathy [21]. The differential production and secretion of these factors lead to non-proliferative and proliferative diabetic retinopathy (i.e. leaky vasculature and neovascularisation) [21]. No change was detected for either factor in the present study. The absence of these two growth factors in our analysis is probably due to technical constraints of the 2-D gels and secretion of these growth factors from the RPE into the extracellular space. For example, the pI of VEGF-A is 8.5, which is outside the pI range of our first dimension (i.e. pI 5–8) and thus would not resolve under our experimental conditions. PEDF was detected in previous 2-D analyses but only in extracellular fractions such as vitreous [22], suggesting that the intracellular content is not sufficient in RPE homogenates for detection in 2-D gels. Consistent with our results, these growth factors were not reported in a previous characterisation of the human RPE cellular proteome [23].
To date, biochemical studies of human diabetic retinopathy have been mostly limited to analysis of the vitreous [22, 24–26]. These analyses have revealed several pathways and factors that provided insight into diabetes-related ocular alterations. For example, by measuring levels in the vitreous, several studies have found an imbalance in angiogenic (i.e. VEGF-A) and anti-angiogenic (i.e. sol flt-1 receptor, PEDF) factors to be present in proliferative diabetic retinopathy [25, 26]. Despite their contributions to an understanding of diabetic retinopathy, several limitations are associated with these studies. First, vitreous protein levels are an indirect, secondary measure of intracellular events occurring in the retina. Second, vitreous collection usually occurs during a surgical procedure at the later stages of retinopathy where early biochemical changes leading to pathogenesis may no longer be evident. Third, the vitreous includes a large amount of albumin and immunoglobulin that may overlap with small spots or less abundant proteins and compromise the utility of 2-D gel analysis. Finally, the cellular origin of many vitreous proteins is unclear and may be non-retinal [27].
There are several advantages associated with the present study. First, we directly analysed retinal proteins from affected human tissue. Second, based on direct clinical evaluation, we excluded donor eyes with characteristic diabetic retinopathy and exclusively studied pre-retinopathic RPE. Third, our proteomic approach enabled simultaneous analysis of over 300 protein spots. Finally, the RPE does not contain disproportionately abundant proteins that could otherwise interfere with the resolution of a large number of protein spots.
Of the proteins identified, 29% (Fig. 3) participate in various metabolic pathways including glycolysis (phosphoglycerate mutase and gamma-enolase), the citric acid cycle (dihydrolipoyl dehydrogenase), lipid metabolism (sterol carrier protein x) and ketolysis (succinyl CoA: 3-ketoacid-coenzyme A transferase 1). In other tissues prone to diabetic complications, such as liver, kidney and both skeletal and heart muscle, energy metabolism is one of the key areas affected [28–32]. However, metabolic changes associated with diabetes are tissue-specific. For example, an increase in glycolysis was reported in skeletal muscle from type 1 and type 2 patients [28, 29], while a reduction was demonstrated in diabetic heart [30, 33] and in streptozotocin-induced diabetic rat retinas after 3 months of disease [34]. Our results indicate that systemic metabolic changes observed in other tissues affected by diabetes also occur in the pre-retinopathic RPE. These tissue-specific changes may result in loss of RPE functions that subsequently impact retinal health and photoreceptor activity.
Chaperones comprised a second major functional category that was altered in diabetic RPE (Fig. 3, Table 2). Chaperones mediate protein folding, defend against protein damage and aggregation due to misfolding, assist with translocation of proteins across intracellular membranes and stabilise unstable protein conformers [35]. Chaperones from multiple subcellular compartments were identified, including mitochondria (GRP75 and elongation factor TU), cytosol (heat shock cognate 71) and the endoplasmic reticulum (protein disulphide-isomerase A3). Protein disulphide-isomerase A3 is a specialised endoplasmic reticulum protein that assists with folding and formation of native disulphide bonds in nascent and unfolded proteins [36]. Specific upregulation of this chaperone suggests elevated unfolded proteins in the endoplasmic reticulum, which is associated with endoplasmic reticulum stress [37]. Numerous studies report the presence of endoplasmic reticulum stress in diabetes. For example, endoplasmic reticulum stress markers such as BiP, GRP94 and CHOP are elevated in the Akita diabetic mouse model [38] and in pancreatic beta cells of type 2 diabetic patients [39].
Aldehyde dehydrogenase 2, a mitochondrial isoform of the aldehyde dehydrogenase family, was increased in our study. It is involved in detoxifying reactive aldehydes produced by lipid peroxidation [40]. Increased content of this protein probably reflects increased lipoxidation products in the diabetic RPE. Lipid peroxidation and oxidation of glycated proteins are two major mechanisms of protein damage resulting from oxidative stress in the diabetic retina [41]. Oxidative stress occurs when the production of oxidants, including reactive oxygen species, exceeds the level of antioxidants in the cell [42]. Numerous studies have shown elevated reactive oxygen species and impaired antioxidant defence in the retina of diabetic animal models and humans, as reviewed [42]. Taken together, elevated levels of aldehyde dehydrogenase 2 and chaperones may be a compensatory response to oxidative stress in the human pre-retinopathic RPE.
Some of the major protein categories that were altered in our study, such as energy metabolism, chaperones and cytoskeletal proteins [43, 44], overlap with findings from other tissues affected by diabetes. However, some proteins altered in the present study are completely novel to diabetes, such as proteins involved in retinoid metabolism (CRALBP), membrane dynamics (annexin A4, A7) and protein transport (selenium binding protein). While the consequences of these changes have yet to be determined, they are consistent with the global effect that diabetes has on multiple cellular processes.
In summary, we compared the human RPE proteome in a pre-retinopathic stage of diabetes to age-matched controls using a high-throughput proteomic approach. Our study demonstrates that significant biochemical changes take place in the RPE prior to clinically evident diabetic retinopathy. It also indicates that changes in metabolic proteins parallel those found in other diabetic tissues. Changes in proteins associated with oxidative and endoplasmic reticulum stress also suggest protein damage in the pre-retinopathic RPE. Given the importance of the RPE in supporting the retinal microenvironment, these alterations may be significant in the early pathogenesis of diabetic retinopathy.
Abbreviations
- AMD:
-
age-related macular degeneration
- 2-D:
-
two-dimensional
- MALDI-TOF:
-
matrix-assisted laser desorption/ionisation time-of-flight
- MOWSE:
-
molecular weight search
- MS/MS:
-
tandem mass spectrometry
- PEDF:
-
pigment epithelium-derived factor
- pI:
-
isoelectric point
- RPE:
-
retinal pigment epithelium
- VEGF:
-
vascular endothelial growth factor
References
International Diabetes Federation (2005) Diabetes Atlas. Available from http://www.eatlas.idf.org/, last accessed January 2008
Klein R, Klein BE, Moss SE, Cruickshanks KJ (1998) The Wisconsin epidemiologic study of diabetic retinopathy: XVII The 14-year incidence and progression of diabetic retinopathy and associated risk factors in type 1 diabetes. Ophthalmology 105:1801–1815
Antonetti DA, Barber AJ, Bronson SK et al (2006) Diabetic retinopathy: seeing beyond glucose-induced microvascular disease. Diabetes 55:2401–2411
Park SH, Park JW, Park SJ et al (2003) Apoptotic death of photoreceptors in the streptozotocin-induced diabetic rat retina. Diabetologia 46:1260–1268
Aizu Y, Oyanagi K, Hu J, Nakagawa H (2002) Degeneration of retinal neuronal processes and pigment epithelium in the early stage of the streptozotocin-diabetic rats. Neuropathology 22:161–170
Phipps JA, Fletcher EL, Vingrys AJ (2004) Paired-flash identification of rod and cone dysfunction in the diabetic rat. Invest Ophthalmol Vis Sci 45:4592–4600
Bensaoula T, Ottlecz A (2001) Biochemical and ultrastructural studies in the neural retina and retinal pigment epithelium of STZ-diabetic rats: effect of captopril. J Ocul Pharmacol Ther 17:573–586
Ramsey DJ, Ripps H, Qian H (2006) An electrophysiological study of retinal function in the diabetic female rat. Invest Ophthalmol Vis Sci 47:5116–5124
Klemp K, Larsen M, Sander B, Vaag A, Brockhoff PB, Lund-Andersen H (2004) Effect of short-term hyperglycemia on multifocal electroretinogram in diabetic patients without retinopathy. Invest Ophthalmol Vis Sci 45:3812–3819
Steinberg RH (1985) Interactions between the retinal pigment epithelium and the neural retina. Doc Ophthalmol 60:327–346
Young TA, Wang H, Munk S et al (2005) Vascular endothelial growth factor expression and secretion by retinal pigment epithelial cells in high glucose and hypoxia is protein kinase C-dependent. Exp Eye Res 80:651–662
Foulds WS (1990) The choroidal circulation and retinal metabolism—an overview. Eye 4:243–248
Quin GG, Len AC, Billson FA, Gillies MC (2007) Proteome map of normal rat retina and comparison with the proteome of diabetic rat retina: new insight in the pathogenesis of diabetic retinopathy. Proteomics 7:2636–2650
Wang YD, Wu JD, Jiang ZL et al (2007) Comparative proteome analysis of neural retinas from type 2 diabetic rats by two-dimensional electrophoresis. Curr Eye Res 32:891–901
Nordgaard CL, Berg KM, Kapphahn RJ et al (2006) Proteomics of the retinal pigment epithelium reveals altered protein expression at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci 47:815–822
Ethen CM, Reilly C, Feng X, Olsen TW, Ferrington DA (2006) The proteome of central and peripheral retina with progression of age-related macular degeneration. Invest Ophthalmol Vis Sci 47:2280–2290
Kapphahn RJ, Ethen CM, Peters EA, Higgins L, Ferrington DA (2003) Modified alpha A crystallin in the retina: altered expression and truncation with aging. Biochemistry 42:15310–15325
Olsen TW, Feng X (2004) The Minnesota Grading System of eye bank eyes for age-related macular degeneration. Invest Ophthalmol Vis Sci 45:4484–4490
Harris WA, Janecki DJ, Reilly JP (2002) Use of matrix clusters and trypsin autolysis fragments as mass calibrants in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 16:1714–1722
Hoppe G, O'Neil J, Hoff HF, Sears J (2004) Products of lipid peroxidation induce missorting of the principal lysosomal protease in retinal pigment epithelium. Biochim Biophys Acta 1689:33–41
Witmer AN, Vrensen GF, Van Noorden CJ, Schlingemann RO (2003) Vascular endothelial growth factors and angiogenesis in eye disease. Prog Retin Eye Res 22:1–29
Yamane K, Minamoto A, Yamashita H et al (2003) Proteome analysis of human vitreous proteins. Mol Cell Proteomics 2:1177–1187
West KA, Yan L, Shadrach K et al (2003) Protein database, human retinal pigment epithelium. Mol Cell Proteomics 2:37–49
Ouchi M, West K, Crabb JW, Kinoshita S, Kamei M (2005) Proteomic analysis of vitreous from diabetic macular edema. Exp Eye Res 81:176–182
Patel JI, Tombran-Tink J, Hykin PG, Gregor ZJ, Cree IA (2006) Vitreous and aqueous concentrations of proangiogenic, antiangiogenic factors and other cytokines in diabetic retinopathy patients with macular edema: implications for structural differences in macular profiles. Exp Eye Res 82:798–806
Nakanishi T, Koyama R, Ikeda T, Shimizu A (2002) Catalogue of soluble proteins in the human vitreous humor: comparison between diabetic retinopathy and macular hole. J Chromatogr B Analyt Technol Biomed Life Sci 776:89–100
Bishop PN, Takanosu M, Le Goff M, Mayne R (2002) The role of the posterior ciliary body in the biosynthesis of vitreous humour. Eye 16:454–460
Crowther GJ, Milstein JM, Jubrias SA, Kushmerick MJ, Gronka RK, Conley KE (2003) Altered energetic properties in skeletal muscle of men with well-controlled insulin-dependent (type 1) diabetes. Am J Physiol Endocrinol Metab 284:E655–E662
Simoneau JA, Kelley DE (1997) Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J Appl Physiol 83:166–171
Stanley WC, Lopaschuk GD, McCormack JG (1997) Regulation of energy substrate metabolism in the diabetic heart. Cardiovasc Res 34:25–33
McLean MP, Billheimer JT, Warden KJ, Irby RB (1995) Differential expression of hepatic sterol carrier proteins in the streptozotocin-treated diabetic rat. Endocrinology 136:3360–3368
Tessari P, Puricelli L, Iori E et al (2007) Altered chaperone and protein turnover regulators expression in cultured skin fibroblasts from type 1 diabetes mellitus with nephropathy. J Proteome Res 6:976–986
Puricelli L, Iori E, Millioni R et al (2006) Proteome analysis of cultured fibroblasts from type 1 diabetic patients and normal subjects. J Clin Endocrinol Metab 91:3507–3514
Ola MS, Berkich DA, Xu Y et al (2006) Analysis of glucose metabolism in diabetic rat retinas. Am J Physiol Endocrinol Metab 290:E1057–E1067
Borges JC, Ramos CH (2005) Protein folding assisted by chaperones. Protein Pept Lett 12:257–261
Yao Y, Zhou Y, Wang C (1997) Both the isomerase and chaperone activities of protein disulfide isomerase are required for the reactivation of reduced and denatured acidic phospholipase A2. Embo J 16:651–658
Yoshida H (2007) ER stress and diseases. Febs J 274:630–658
Nozaki J, Kubota H, Yoshida H et al (2004) The endoplasmic reticulum stress response is stimulated through the continuous activation of transcription factors ATF6 and XBP1 in Ins2+/Akita pancreatic beta cells. Genes Cells 9:261–270
Laybutt DR, Preston AM, Akerfeldt MC et al (2007) Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia 50:752–763
Ohta S, Ohsawa I (2006) Dysfunction of mitochondria and oxidative stress in the pathogenesis of Alzheimer’s disease: on defects in the cytochrome c oxidase complex and aldehyde detoxification. J Alzheimers Dis 9:155–166
van Reyk DM, Gillies MC, Davies MJ (2003) The retina: oxidative stress and diabetes. Redox Rep 8:187–192
Ellis EM (2007) Reactive carbonyls and oxidative stress: potential for therapeutic intervention. Pharmacol Ther 115:13–24
Dhalla NS, Liu X, Panagia V, Takeda N (1998) Subcellular remodeling and heart dysfunction in chronic diabetes. Cardiovasc Res 40:239–247
McLean WG (1997) The role of axonal cytoskeleton in diabetic neuropathy. Neurochem Res 22:951–956
Murata C, Taniyama M, Kuriyama S et al (2004) Meta-analysis of three diabetes population studies: association of inactive ALDH2 genotype with maternal inheritance of diabetes. Diabetes Res Clin Pract 66(Suppl 1):S145–S147
Wadham C, Parker A, Wang L, Xia P (2007) High glucose attenuates protein S-nitrosylation in endothelial cells: role of oxidative stress. Diabetes 56:2715–2721
Witek B, Krol T, Kolataj A, Ochwanowska E, Stanislawska I, Slewa A (2001) The insulin, glucose and cholesterol level and activity of lysosomal enzymes in the course of the model alloxan diabetes. Neuro Endocrinol Lett 22:238–242
Sreekumar R, Halvatsiotis P, Schimke JC, Nair KS (2002) Gene expression profile in skeletal muscle of type 2 diabetes and the effect of insulin treatment. Diabetes 51:1913–1920
Yokoyama T, Yamane K, Minamoto A et al (2006) High glucose concentration induces elevated expression of anti-oxidant and proteolytic enzymes in cultured human retinal pigment epithelial cells. Exp Eye Res 83:602–609
Turko IV, Marcondes S, Murad F (2001) Diabetes-associated nitration of tyrosine and inactivation of succinyl-CoA:3-oxoacid CoA-transferase. Am J Physiol Heart Circ Physiol 281:H2289–H2294
Acknowledgements
This research was supported in part by grants EY014176 (to D. A. Ferrington) and AG025392 (to T. W. Olsen) from the National Institutes of Health, by the Minnesota Lions Macular Degeneration Center, by a career development award from the American Federation for Aging Research and Foundation Fighting Blindness (to D. A. Ferrington), by the Minnesota Medical Foundation and by an unrestricted grant from Research to Prevent Blindness Foundation. The authors thank the Minnesota Lions Eye Bank for their assistance in procuring eyes for this study. The mass spectrometry analysis was performed at the Mass Spectrometry Consortium for the Life Sciences at the University Minnesota.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. Decanini and P. R. Karunadharma contributed equally to this study.
Rights and permissions
About this article
Cite this article
Decanini, A., Karunadharma, P.R., Nordgaard, C.L. et al. Human retinal pigment epithelium proteome changes in early diabetes. Diabetologia 51, 1051–1061 (2008). https://doi.org/10.1007/s00125-008-0991-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-008-0991-2