Abstract
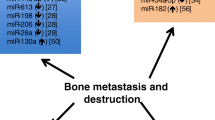
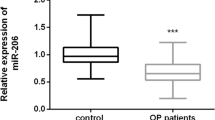
Starting from their role exerted on osteoblast and osteoclast differentiation and activity pathways, microRNAs (miRNAs) have been recently identified as regulators of different processes in bone homeostasis. For this purpose, in a recent review, we highlighted, as deregulated miRNAs could be involved in different bone diseases such as osteoporosis. In addition, recent studies supported the concept that osteoporosis-induced bone alterations might offer a receptive site for cancer cells to form bone metastases, However, to date, no data on specific-shared miRNAs between osteoporosis and bone metastases have been considered and described to clarify the evidence of this link. The main goal of this review is to underline as deregulated miRNAs in osteoporosis may have specific roles in the development of bone metastases. The review showed that several circulating osteoporotic miRNAs could facilitate tumor progression and bone-metastasis formation in several tumor types, i.e., breast cancer, prostate cancer, non-small-cell lung cancer, esophageal squamous cell carcinoma, and multiple myeloma. In detail, serum up-regulation of pro-osteoporotic miRNAs, as well as serum down-regulation of anti-osteoporotic miRNAs are common features of all these tumors and are able to promote bone metastasis. These results are of key importance and could help researcher and clinicians to establish new therapeutic strategies connected with deregulation of circulating miRNAs and able to interfere with pathogenic processes of osteoporosis, tumor progressions, and bone-metastasis formation.



Similar content being viewed by others
Abbreviations
- 3′UTR:
-
3′ Untranslated region
- AGO2:
-
Argonaut 2
- BC:
-
Breast cancer
- BMSCs:
-
Bone-marrow mesenchymal stem cells
- BMPs:
-
Bone morphogenetic proteins
- CNTF:
-
Ciliary neurotrophic factor
- DGCR8:
-
Di George syndrome critical region gene 8
- ESCC:
-
Esophageal squamous cell carcinoma
- GPC3:
-
Glypican 3
- IL:
-
Interleukin
- MM:
-
Multiple myeloma
- MMP:
-
Matrix metalloproteinase
- MTSS1:
-
Metastasis suppressor protein 1
- MiRNAs:
-
MicroRNAs
- NSCLC:
-
Non-small-cell lung cancer
- OB:
-
Osteoblast
- OC:
-
Osteoclast
- OPG:
-
Osteoprotegerin
- PC:
-
Prostate cancer
- PTH:
-
Parathyroid hormone
- PBMCs:
-
Peripheral blood mononuclear cells
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- RANKL:
-
Receptor activator of nuclear factor kappa-B ligand
- SREs:
-
Skeletal-related events
- TGFβ:
-
Transforming growth factor-beta
- TNFα:
-
Tumor necrosis factor alpha
- Wnt:
-
Wingless-type MMTV integration site family
References
Florencio-Silva R, Sasso GR, Sasso-Cerri E, Simões MJ, Cerri PS (2015) Biology of bone tissue: structure, function, and factors that influence bone cells. Biomed Res Int 2015:421746
Komori T (2011) Signaling networks in RUNX2-dependent bone development. J Cell Biochem 112:750–755
Guntur AR, Rosen CJ (2013) IGF-1 regulation of key signaling pathways in bone. Bonekey Rep 2:437
Wu M, Chen G, Li YP (2016) TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res 4:16009
Larousserie F et al (2017) Frontline science: human bone cells as a source of IL-27 under inflammatory conditions: role of TLRs and cytokines. J Leukoc Biol 101:1289–1300
Yamada A et al (2007) Interleukin-4 inhibition of osteoclast differentiation is stronger than that of interleukin-13 and they are equivalent for induction of osteoprotegerin production from osteoblasts. Immunology 120:573–579
Silfverswärd CJ, Penno H, Frost A, Nilsson O, Ljunggren O (2010) Expression of markers of activity in cultured human osteoblasts: effects of interleukin-4 and interleukin-13. Scand J Clin Lab Investig 70:338–342
Forte GI et al (2009) Characterization of two alternative Interleukin(IL)-10 5′UTR mRNA sequences, induced by lipopolysaccharide (LPS) stimulation of peripheral blood mononuclear cells. Mol Immunol 46:2161–2166
Zhang Q, Chen B, Yan F, Guo J, Zhu X, Ma S, Yang W (2014) Interleukin-10 inhibits bone resorption: a potential therapeutic strategy in periodontitis and other bone loss diseases. Biomed Res Int 2014:284836
Bellavia D, Costa V, De Luca A, Maglio M, Pagani S, Fini M, Giavaresi G (2016) Vitamin D level between calcium-phosphorus homeostasis and immune system: new perspective in osteoporosis. Curr Osteoporos Rep. https://doi.org/10.1007/s11914-016-0331-2
Kraemer B, Rothmund R, Banys M, Krawczyk N, Solomayer EF, Mack C, Wallwiener D, Fehm T (2011) Impaired bone microenvironment: correlation between bone density and presence of disseminated tumor cells. Anticancer Res 31:4423–4428
Coleman R et al (2014) Adjuvant zoledronic acid in patients with early breast cancer: final efficacy analysis of the AZURE (BIG 01/04) randomised open-label phase 3 trial. Lancet Oncol 15:997–1006
Ottewell PD, Wang N, Brown HK, Reeves KJ, Fowles CA, Croucher PI, Eaton CL, Holen I (2014) Zoledronic acid has differential antitumor activity in the pre- and postmenopausal bone microenvironment in vivo. Clin Cancer Res 20:2922–2932
Pagani S, Fini M, Giavaresi G, Salamanna F, Borsari V (2015) The active role of osteoporosis in the interaction between osteoblasts and bone metastases. Bone 79:176–182
Salamanna F et al (2016) An in vitro 3D bone metastasis model by using a human bone tissue culture and human sex-related cancer cells. Oncotarget 7:76966–76983
Salamanna F, Contartese D, Maglio M, Fini M (2016) A systematic review on in vitro 3D bone metastases models: a new horizon to recapitulate the native clinical scenario? Oncotarget 7:44803–44820
Salamanna F, Pagani S, Maglio M, Borsari V, Giavaresi G, Martelli AM, Buontempo F, Fini M (2016) Estrogen-deficient osteoporosis enhances the recruitment and activity of osteoclasts by breast cancer cells. Histol Histopathol 31:83–93
Salamanna F, Borsari V, Brogini S, Torricelli P, Cepollaro S, Cadossi M, Fini M (2017) A human 3D in vitro model to assess the relationship between osteoporosis and dissemination to bone of breast cancer tumor cells. J Cell Physiol 232:1826–1834
Salamanna F, Borsari V, Contartese D, Nicoli Aldini N, Fini M (2018) Link between estrogen deficiency osteoporosis and susceptibility to bone metastases: a way towards precision medicine in cancer patients. Breast 41:42–50
Coleman RE (2006) Clinical features of metastatic bone disease and risk of skeletal morbidity. Clin Cancer Res 12:6243s–6249s
David Roodman G, Silbermann R (2015) Mechanisms of osteolytic and osteoblastic skeletal lesions. Bonekey Rep 4:753
Landgraf P et al (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414
Mizoguchi F et al (2010) Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem 109:866–875
Bartel DP (2018) Metazoan microRNAs. Cell 173:20–51
Bellavia D et al (2019) Deregulated miRNAs in bone health: epigenetic roles in osteoporosis. Bone 122:52–75. https://doi.org/10.1016/j.bone.2019.02.013
Li H et al (2009) A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Investig 119:3666–3677
Wang Y, Li L, Moore BT, Peng XH, Fang X, Lappe JM, Recker RR, Xiao P (2012) MiR-133a in human circulating monocytes: a potential biomarker associated with postmenopausal osteoporosis. PLoS One 7:e34641
Wang X et al (2013) miR-214 targets ATF4 to inhibit bone formation. Nat Med 19:93–100
Yang N et al (2013) Tumor necrosis factor α suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J Bone Miner Res 28:559–573
Chen S et al (2014) MicroRNA-125b suppresses the proliferation and osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Mol Med Rep 9:1820–1826
Seeliger C, Karpinski K, Haug AT, Vester H, Schmitt A, Bauer JS, van Griensven M (2014) Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res 29:1718–1728
Li H, Wang Z, Fu Q, Zhang J (2014) Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers 19:553–556
Chen C, Cheng P, Xie H, Zhou HD, Wu XP, Liao EY, Luo XH (2014) MiR-503 regulates osteoclastogenesis via targeting RANK. J Bone Miner Res 29:338–347
Cao Z, Moore BT, Wang Y, Peng XH, Lappe JM, Recker RR, Xiao P (2014) MiR-422a as a potential cellular microRNA biomarker for postmenopausal osteoporosis. PLoS One 9:e97098
Krzeszinski JY et al (2014) miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 512:431–435
Lv H, Sun Y, Zhang Y (2015) MiR-133 is involved in estrogen deficiency-induced osteoporosis through modulating osteogenic differentiation of mesenchymal stem cells. Med Sci Monit 21:1527–1534
Weilner S et al (2015) Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone 79:43–51
Garmilla-Ezquerra P, Sañudo C, Delgado-Calle J, Pérez-Nuñez MI, Sumillera M, Riancho JA (2015) Analysis of the bone microRNome in osteoporotic fractures. Calcif Tissue Int 96:30–37
De-Ugarte L et al (2015) MiRNA profiling of whole trabecular bone: identification of osteoporosis-related changes in MiRNAs in human hip bones. BMC Med Genom 8:75
Kocijan R et al (2016) Circulating microRNA signatures in patients with idiopathic and postmenopausal osteoporosis and fragility fractures. J Clin Endocrinol Metab 101:4125–4134
Bedene A et al (2016) MiR-148a the epigenetic regulator of bone homeostasis is increased in plasma of osteoporotic postmenopausal women. Wien Klin Wochenschr 128:519–526
You L, Pan L, Chen L, Gu W, Chen J (2016) MiR-27a is essential for the shift from osteogenic differentiation to adipogenic differentiation of mesenchymal stem cells in postmenopausal osteoporosis. Cell Physiol Biochem 39:253–265
Chen J et al (2016) Identification of suitable reference gene and biomarkers of serum miRNAs for osteoporosis. Sci Rep 6:36347
Wang Q et al (2017) LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed Pharmacother 89:1178–1186
Yavropoulou MP, Anastasilakis AD, Makras P, Tsalikakis DG, Grammatiki M, Yovos JG (2017) Expression of microRNAs that regulate bone turnover in the serum of postmenopausal women with low bone mass and vertebral fractures. Eur J Endocrinol 176:169–176
Kelch S, Balmayor ER, Seeliger C, Vester H, Kirschke JS, van Griensven M (2017) miRNAs in bone tissue correlate to bone mineral density and circulating miRNAs are gender independent in osteoporotic patients. Sci Rep 7:15861
Jiménez-Ortega RF et al (2017) Identification of microRNAs in human circulating monocytes of postmenopausal osteoporotic Mexican-Mestizo women: a pilot study. Exp Ther Med 14:5464–5472
Chen YJ, Chang WA, Huang MS, Chen CH, Wang KY, Hsu YL, Kuo PL (2017) Identification of novel genes in aging osteoblasts using next-generation sequencing and bioinformatics. Oncotarget 8:113598–113613
Hu CH, Sui BD, Du FY, Shuai Y, Zheng CX, Zhao P, Yu XR, Jin Y (2017) miR-21 deficiency inhibits osteoclast function and prevents bone loss in mice. Sci Rep 7:43191
Feichtinger X et al (2018) Bone-related circulating microRNAs miR-29b-3p, miR-550a-3p, and miR-324-3p and their association to bone microstructure and histomorphometry. Sci Rep 8:4867
Liu H, Liu Q, Wu XP, He HB, Fu L (2018) MiR-96 regulates bone metabolism by targeting osterix. Clin Exp Pharmacol Physiol 45:602–613
Ren H et al (2018) miRNA-seq analysis of human vertebrae provides insight into the mechanism underlying GIOP. Bone 120:371–386
Wang C, He H, Wang L, Jiang Y, Xu Y (2018) Reduced miR-144-3p expression in serum and bone mediates osteoporosis pathogenesis by targeting RANK. Biochem Cell Biol 96:627–635
Ramírez-Salazar EG et al (2018) Serum miRNAs miR-140-3p and miR-23b-3p as potential biomarkers for osteoporosis and osteoporotic fracture in postmenopausal Mexican-Mestizo women. Gene 679:19–27
Aquino-Martinez R et al (2018) miR-219a-5p regulates Rorβ during osteoblast differentiation and in age-related bone loss. J Bone Miner Res 34:135–144. https://doi.org/10.1002/jbmr.3586
Mencía Castaño I, Curtin CM, Duffy GP, O'Brien FJ (2018) Harnessing a novel inhibitory role of miR-16 in osteogenesis by human mesenchymal stem cells for advanced scaffold-based bone tissue engineering. Tissue Eng Part A 25:24–33. https://doi.org/10.1089/ten.TEA.2017.0460
Mäkitie RE, Hackl M, Niinimäki R, Kakko S, Grillari J, Mäkitie O (2018) Altered microRNA profile in osteoporosis caused by impaired WNT signaling. J Clin Endocrinol Metab 103:1985–1996
Haider MT, Taipaleenmäki H (2018) Targeting the metastatic bone microenvironment by MicroRNAs. Front Endocrinol (Lausanne) 9:202
Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K, Amadori D, Kang Y (2013) Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell 24:542–556
Stückrath I, Rack B, Janni W, Jäger B, Pantel K, Schwarzenbach H (2015) Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget 6:13387–13401
Wang F, Zheng Z, Guo J, Ding X (2010) Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol 119:586–593
Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS (2011) Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem 57:84–91
Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodríguez-Padilla C, Reséndez-Pérez D (2013) Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers 34:163–169
Matamala N et al (2015) Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem 61:1098–1106
Liu X, Feng J, Tang L, Liao L, Xu Q, Zhu S (2015) The regulation and function of miR-21-FOXO3a-miR-34b/c signaling in breast cancer. Int J Mol Sci 16:3148–3162
Wang G, Wang L, Sun S, Wu J, Wang Q (2015) Quantitative measurement of serum microRNA-21 expression in relation to breast cancer metastasis in Chinese females. Ann Lab Med 35:226–232
Jurkovicova D et al (2016) Evaluation of expression profiles of microRNAs and two target genes, FOXO3a and RUNX2, effectively supports diagnostics and therapy predictions in breast cancer. Neoplasma 63:941–951
Liu B et al (2017) Serum miR-21 and miR-125b as markers predicting neoadjuvant chemotherapy response and prognosis in stage II/III breast cancer. Hum Pathol 64:44–52
Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, Hu C (2013) Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol 139:223–229
Wang H, Tan G, Dong L, Cheng L, Li K, Wang Z, Luo H (2012) Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One 7:e34210
Chan M et al (2013) Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res 19:4477–4487
Liu J et al (2017) Osteoclastic miR-214 targets TRAF3 to contribute to osteolytic bone metastasis of breast cancer. Sci Rep 7:40487
Schwarzenbach H, Milde-Langosch K, Steinbach B, Müller V, Pantel K (2012) Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res Treat 134:933–941
Maroni P, Puglisi R, Mattia G, Carè A, Matteucci E, Bendinelli P, Desiderio MA (2017) In bone metastasis miR-34a-5p absence inversely correlates with Met expression, while Met oncogene is unaffected by miR-34a-5p in non-metastatic and metastatic breast carcinomas. Carcinogenesis 38:492–503
Zeng Z, Chen X, Zhu D, Luo Z, Yang M (2017) Low expression of circulating microRNA-34c is associated with poor prognosis in triple-negative breast cancer. Yonsei Med J 58:697–702
Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449:682–688
Zhao FL, Hu GD, Wang XF, Zhang XH, Zhang YK, Yu ZS (2012) Serum overexpression of microRNA-10b in patients with bone metastatic primary breast cancer. J Int Med Res 40:859–866
Bašová P, Pešta M, Sochor M, Stopka T (2017) Prediction potential of Serum miR-155 and miR-24 for relapsing early breast cancer. Int J Mol Sci 18:1–10. https://doi.org/10.3390/ijms18102116
Croset M et al (2018) miRNA-30 Family members inhibit breast cancer invasion, osteomimicry, and bone destruction by directly targeting multiple bone metastasis-associated genes. Cancer Res 78:5259–5273
Kong X, Zhang J, Li J, Shao J, Fang L (2018) MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem Biophys Res Commun 501:486–493
Madhavan D et al (2016) Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis 37:461–470
van Schooneveld E et al (2012) Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res 14:R34
Ng EK et al (2013) Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One 8:e53141
Lin SC, Yu-Lee LY, Lin SH (2018) Osteoblastic factors in prostate cancer bone metastasis. Curr Osteoporos Rep 16:642–647
Guo X, Han T, Hu P, Zhu C, Wang Y, Chang S (2018) Five microRNAs in serum as potential biomarkers for prostate cancer risk assessment and therapeutic intervention. Int Urol Nephrol 50:2193–2200
Zhang HL et al (2011) Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 71:326–331
Watahiki A, Macfarlane RJ, Gleave ME, Crea F, Wang Y, Helgason CD, Chi KN (2013) Plasma miRNAs as biomarkers to identify patients with castration-resistant metastatic prostate cancer. Int J Mol Sci 14:7757–7770
Bonci D et al (2016) A microRNA code for prostate cancer metastasis. Oncogene 35:1180–1192
Chen WY et al (2015) MicroRNA-34a regulates WNT/TCF7 signaling and inhibits bone metastasis in Ras-activated prostate cancer. Oncotarget 6:441–457
Fang Y, Qiu J, Jiang ZB, Xu SR, Zhou ZH, He RL (2019) Increased serum levels of miR-214 in patients with PCa with bone metastasis may serve as a potential biomarker by targeting PTEN. Oncol Lett 17:398–405
Nishikawa R et al (2014) Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int J Oncol 45:401–410
Zhu C, Hou X, Zhu J, Jiang C, Wei W (2018) Expression of miR-30c and miR-29b in prostate cancer and its diagnostic significance. Oncol Lett 16:3140–3144
Xu L, Zhong J, Guo B, Zhu Q, Liang H, Wen N, Yun W, Zhang L (2016) miR-96 promotes the growth of prostate carcinoma cells by suppressing MTSS1. Tumour Biol 37:12023–12032
Tsai YC, Chen WY, Siu MK, Tsai HY, Yin JJ, Huang J, Liu YN (2017) Epidermal growth factor receptor signaling promotes metastatic prostate cancer through microRNA-96-mediated downregulation of the tumor suppressor ETV6. Cancer Lett 384:1–8
Siu MK, Tsai YC, Chang YS, Yin JJ, Suau F, Chen WY, Liu YN (2015) Transforming growth factor-β promotes prostate bone metastasis through induction of microRNA-96 and activation of the mTOR pathway. Oncogene 34:4767–4776
Singh PK et al (2014) Serum microRNA expression patterns that predict early treatment failure in prostate cancer patients. Oncotarget 5:824–840
Wei J, Gao W, Zhu CJ, Liu YQ, Mei Z, Cheng T, Shu YQ (2011) Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer 30:407–414
Shen J et al (2011) Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer 11:374
Zhao K, Cheng J, Chen B, Liu Q, Xu D, Zhang Y (2017) Circulating microRNA-34 family low expression correlates with poor prognosis in patients with non-small cell lung cancer. J Thorac Dis 9:3735–3746
Wang C et al (2015) A five-miRNA panel identified from a multicentric case-control study serves as a novel diagnostic tool for ethnically diverse non-small-cell lung cancer patients. EBioMedicine 2:1377–1385
Liu Y, Zhang G, Li H, Han L, Fu A, Zhang N, Zheng Y (2015) Serum microRNA-365 in combination with its target gene TTF-1 as a non-invasive prognostic marker for non-small cell lung cancer. Biomed Pharmacother 75:185–190
Silva J et al (2011) Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J 37:617–623
Zhu W, Liu X, He J, Chen D, Hunag Y, Zhang YK (2011) Overexpression of members of the microRNA-183 family is a risk factor for lung cancer: a case control study. BMC Cancer 11:393
Fei X, Zhang J, Zhao Y, Sun M, Zhao H, Li S (2018) miR-96 promotes invasion and metastasis by targeting GPC3 in non-small cell lung cancer cells. Oncol Lett 15:9081–9086
Wang T et al (2012) Cell-free microRNA expression profiles in malignant effusion associated with patient survival in non-small cell lung cancer. PLoS One 7:e43268
Yuxia M, Zhennan T, Wei Z (2012) Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J Cancer Res Clin Oncol 138:2045–2050
Cui EH, Li HJ, Hua F, Wang B, Mao W, Feng XR, Li JY, Wang X (2013) Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol Sin 34:309–313
Chen X et al (2012) Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer 130:1620–1628
Zhang C et al (2010) Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem 56:1871–1879
Wu C et al (2014) Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One 9:e92292
Wu C, Li M, Hu C, Duan H (2014) Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep 41:1257–1266
Khazaei S, Nouraee N, Moradi A, Mowla SJ (2017) A novel signaling role for miR-451 in esophageal tumor microenvironment and its contribution to tumor progression. Clin Transl Oncol 19:633–640
Dong W, Li B, Wang J, Song Y, Zhang Z, Fu C, Zhang P (2016) Diagnostic and predictive significance of serum microRNA-7 in esophageal squamous cell carcinoma. Oncol Rep 35:1449–1456
Lin X, Xu XY, Chen QS, Huang C (2015) Clinical significance of microRNA-34a in esophageal squamous cell carcinoma. Genet Mol Res 14:17684–17691
Cui XB et al (2017) MicroRNA-34a functions as a tumor suppressor by directly targeting oncogenic PLCE1 in Kazakh esophageal squamous cell carcinoma. Oncotarget 8:92454–92469
Dong W, Li B, Wang Z, Zhang Z, Wang J (2015) Clinical significance of microRNA-24 expression in esophageal squamous cell carcinoma. Neoplasma 62:250–258
Lu YF et al (2018) Promoter hypomethylation mediated upregulation of MicroRNA-10b-3p targets FOXO3 to promote the progression of esophageal squamous cell carcinoma (ESCC). J Exp Clin Cancer Res 37:301
Wang K, Chen D, Meng Y, Xu J, Zhang Q (2018) Clinical evaluation of 4 types of microRNA in serum as biomarkers of esophageal squamous cell carcinoma. Oncol Lett 16:1196–1204
Raimondi L et al (2015) Involvement of multiple myeloma cell-derived exosomes in osteoclast differentiation. Oncotarget 6:13772–13789
Wang W et al (2015) Aberrant levels of miRNAs in bone marrow microenvironment and peripheral blood of myeloma patients and disease progression. J Mol Diagn 17:669–678
Hao M et al (2016) Serum high expression of miR-214 and miR-135b as novel predictor for myeloma bone disease development and prognosis. Oncotarget 7:19589–19600
Zhang YK et al (2011) Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1. Biochem Biophys Res Commun 414:233–239
Bellavia D et al (2018) Engineered exosomes: a new promise for the management of musculoskeletal diseases. Biochim Biophys Acta 1862:1893–1901
Acknowledgements
Dr. Daniele Bellavia, Dr. Angela De Luca, Dr. Valeria Carina, Dr. Viviana Costa, and Dr. Lavinia Raimondi contributed to the manuscript by working at the Technology Platform of Tissue Engineering, Theranostics and Oncology (Lab.Manager Dr. Gianluca Giavaresi), a laboratory started-up by the Rizzoli Orthopedic Institute in Palermo (Italy) with the grants also of National Operative Program projects (PON, MIUR).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests. No benefits in any form were received or will be received from a commercial party related directly or indirectly to the subject of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bellavia, D., Salamanna, F., Raimondi, L. et al. Deregulated miRNAs in osteoporosis: effects in bone metastasis. Cell. Mol. Life Sci. 76, 3723–3744 (2019). https://doi.org/10.1007/s00018-019-03162-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-019-03162-w