Abstract
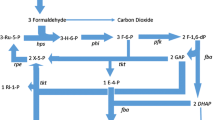
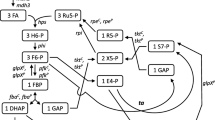
The enzymology of methanol utilization in thermotolerant methylotrophic Bacillus strains was investigated. In all strains an immunologically related NAD-dependent methanol dehydrogenase was involved in the initial oxidation of methanol. In cells of Bacillus sp. C1 grown under methanol-limiting conditions this enzyme constituted a high percentage of total soluble protein. The methanol dehydrogenase from this organism was purified to homogeneity and characterized. In cell-free extracts the enzyme displayed biphasic kinetics towards methanol, with apparent K m values of 3.8 and 166 mM. Carbon assimilation was by way of the fructose-1,6-bisphosphate aldolase cleavage and transketolase/transaldolase rearrangement variant of the RuMP cycle of formaldehyde fixation. The key enzymes of the RuMP cycle, hexulose-6-phosphate synthase (HPS) and hexulose-6-phosphate isomerase (HPI), were present at very high levels of activity. Failure of whole cells to oxidize formate, and the absence of formaldehyde-and formate dehydrogenases indicated the operation of a non-linear oxidation sequence for formaldehyde via HPS. A comparison of the levels of methanol dehydrogenase and HPS in cells of Bacillus sp. C1 grown on methanol and glucose suggested that the synthesis of these enzymes is not under coordinate control.
Similar content being viewed by others
Abbreviations
- RuMP:
-
ribulose monophosphate
- HPS:
-
hexulose-6-phosphate synthase
- HPI:
-
hexulose-6-phosphate isomerase
- MDH:
-
methanol dehydrogenase
- ADH:
-
acohol dehydrogenase
- PQQ:
-
pyrroloquinoline, quinone
- DTT:
-
dithiothreitol
- NBT:
-
nitrobluetetrazolium
- PMS:
-
phenazine methosulphate
- DCPIP:
-
dichlorophenol indophenol
References
Al-Awadhi N, Egli T, Hamer G (1988) Growth characteristics of a thermotolerant methylotrophic Bacillus sp. (NCIB 12522) in batch culture. Appl Microbiol Biotechnol 29:485–493
Anthony C (1982) The biochemistry of methylotrophs. Academic Press, London
Anthony C (1986) Bacterial oxidation of methane and methanol. Adv Microb Physiol 27:113–210
Attwood MM, Quayle JR (1984) Formaldehyde as a central metabolite of methylotrophic metabolism. In: Crawford RL, Hanson RS (eds) Microbial growth on C1 compounds. American Society for Microbiology, Washington DC, pp 315–323
Avigad G (1983) A simple spectrophotometric determination of formaldehyde and other aldehydes: Application to periodateoxidized glycol systems. Anal Biochem 134:499–504
Bellion E, Wu GTS (1978) Alcohol dehydrogenases from a facultative methylotrophic bacterium. J Bacteriol 135:252–258
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brändén CI, Jörnvall H, Eklund H, Furugren B (1975) Alcohol dehydrogenases. In: Boyer PD (ed) The enzymes, 3rd edn, vol 11 A. Academic Press, New York, pp 103–190
Brooke AG, Attwood MM (1983) Regulation of enzyme synthesis during growth of Hyphomicrobium X on mixtures of methylamine and ethanol. J Gen Microbiol 129:2399–2404
Croes GM, Levering PR, Dijkhuizen L (1986) Regulation of methanol metabolism and carbon dioxide fixation in Xanthobacter autotrophicus strain 25a. Abstr 5th Int Symp on Microbial Growth on C1 Compounds. Haren. Free University Press, Amsterdam, p 13
Dijken JP van, Quayle JR (1977) Fructose metabolism in four Pseudomonas species. Arch Microbiol 114:281–286
Dijken JP van, Oostra-Demkes GJ, Otto R, Harder W (1976) S-Formylglutathione: the substrate for formate dehydrogenase in methanol-utilizing yeasts. Arch Microbiol 111:77–84
Dijken JP van, Harder W, Beardsmore AJ, Quayle JR (1978) Dihydroxyacetone: an intermediate in the assimilation of methanol by yeasts? FEMS Microbiol Lett 4:97–102
Dijkhuizen L, Knight M, Harder W (1978) Metabolic regulation in Pseudomonas oxalaticus OX1. Autotrophic and heterotrophic growth on mixed substrates. Arch Microbiol 116:77–83
Dijkhuizen L, Levering PR (1987) Metabolic regulation in facultative methylotrophs. In: Verseveld HW van, Duine JA (eds) Microbial growth on C1 compounds. Nijhoff, Dordrecht, pp 95–104
Dijkhuizen L, Arfman N, Attwood MM, Brooke AG, Harder W, Watling EM (1988) Isolation and initial characterization of thermotolerant methylotrophic Bacillus strains. FEMS Microbiol Lett 52:209–214
Douma AC, Veenhuis M, de Koning W, Evers M, Harder W (1985) Dihydroxyacetone synthase is localized in the peroxisomal matrix of methanol-grown Hansenula polymorpha. Arch Microbiol 143:237–243
Dowds BCA, Sheehan MC, Bailey CJ, McConnell DJ (1988) Cloning and characterization of the gene for a methanolutilising alcohol dehydrogenase from Bacillus stearothermophilus. Gene 68:11–22
Duester G, Jornvall H, Hatfield GW (1986) Intron-dependent evolution of the nucleotide-binding domains within alcohol dehydrogenase and related enzymes. Nucleic Acids Res 14:1931–1941
Duine JA, Frank J, Westerling J (1978) Purification and properties of methanol dehydrogenase from Hyphomicrobium X. Biochim Biophys Acta 524:277–287
Duine JA, Frank J, Berkhout MPJ (1984) NAD-dependent, PQQ-containing methanol dehydrogenase: a bacterial dehydrogenase in a multienzyme complex. FEBS Lett 168:217–221
Duine JA, Frank J, Jongejan JA (1986) PQQ and quinoprotein enzymes in microbial oxidations. FEMS Microbiol Rev 32:165–178
Ghosh R, Quayle JR (1981) Purification and properties of the methanol dehydrogenase from Methylophilus methylotrophus. Biochem J 199:245–250
Groen B, Kleef MAG van, Duine JA (1986) Quinohaemoprotein alcohol dehydrogenase apoenzyme from Pseudomonas testosteroni. Biochem J 234:611–615
Hazeu W, Bruyn JC de, Dijken JP van (1983) Nocardia sp. 239, a facultative methanol utilizer with the ribulose monophosphate pathway of formaldehyde fixation. Arch Microbiol 135:205–210
Janssen DB, Keuning S, Witholt B (1987) Involvement of a quinoprotein alcohol dehydrogenase and an NAD-dependent aldehyde dehydrogenase in 2-chloroethanol metabolism in Xanthobacter autotrophicus GJ10. J Gen Microbiol 133:85–92
Leammli UK, Favre K (1973) Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol 80:575–599
Levering PR, Dijken JP van, Veenhuis M, Harder W (1981) Arthrobacter P1, a fast growing versatile methylotroph with amine oxidase as a key enzyme in the metabolism of methylated amines. Arch Microbiol 129:72–80
Levering PR, Dijkhuizen L, Harder W (1982) Enzymatic evidence for the operation of the FBP aldolase cleavage and TK/TA rearrangement varint of the RuMP cycle in Arthrobacter P1. FEMS Microbiol Lett 14:257–261
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Page MD, Anthony C (1986) Regulation of formaldehyde oxidation by the methanol dehydrogenase modifier proteins of Methylophilus methylotrophus and Pseudomonas AM1. J Gen Microbiol 132:1553–1563
Rella R, Raia CA, Pensa M, Pisani FM, Gambacorta A, De Rosa M, Rossi M (1987) A novel archaebacterial NAD+-dependent alcohol dehydrogenase. Purification and propeties. Eur J Biochem 167:475–479
Sheehan MC, Bailey CJ, Dowds BCA, McConnell DJ (1988) A new alcohol dehydrogenase, reactive towards methanol, from Bacillus stearothermophilus. Biochem J 252:661–666
Srere PA (1987) Complexes of sequential metabolic enzymes. Annu Rev Biochem 56:89–124
Steinbüchel A, Schlegel HG (1984) A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: purification and properties. Eur J Biochem 141:555–564
Sund H, Theorell H (1963) Alcohol dehydrogenases. In: Boyer PD, Lardy H, Myrbäck K (eds) The enzymes, 2nd edn, vol 7A. Academic Press, New York, pp 25–83
Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA 76:4350–4354
Verduyn C, Breedveld GJ, Scheffers WA, Dijken JP van (1988) Substrate specificity of alcohol dehydrogenase from the yeasts Hansenula polymorpha CBS 4732 and Candida utilis CBS 621. Yeast 4:143–148
Vries GE de (1986) Molecular biology of bacterial methanol oxidation. FEMS Microbiol Rev 39:235–258
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Arfman, N., Watling, E.M., Clement, W. et al. Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch. Microbiol. 152, 280–288 (1989). https://doi.org/10.1007/BF00409664
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00409664