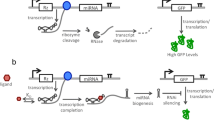
Abstract
Engineering synthetic gene circuits to control cellular functions has a broad application in the field of synthetic biology. Synthetic RNA-based switches that can operate at the transcriptional and posttranscriptional level have also drawn significant interest for the application of next-generation therapeutics and diagnostics. Thus, RNA-based switchable platforms are needed to report dynamic cellular mechanisms which play an important role in cell development and diseases. Recently, several RNA-based switches have been designed and utilized for biosensing and molecular diagnostics. However, miRNA-based switches have not been well established or characterized, especially for eukaryotic translational control. Here, we designed a novel synthetic toehold switch for detection of exogenously and endogenously expressed miRNAs in CHO, HeLa, HEK 293, and MDA-MB-231 breast cancer cells. Multiplex detection of miR-155 and miR-21 was tested using two toehold switches to evaluate the orthogonality and programmability of this synthetic platform.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15:321–333
Hayes J, Peruzzi PP, Lawler S (2014) MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 20:460–469
Jansson MD, Lund AH (2012) MicroRNA and cancer. Mol Oncol 6:590–610
Suzuki HI, Katsura A et al (2015) MicroRNA regulons in tumor microenvironment. Oncogene 34:3085–3094
Forterre A, Komuro H et al (2020) A comprehensive review of cancer microRNA therapeutic delivery strategies. Cancers 12:1852
Wilczynska A, Bushell M (2015) The complexity of miRNA-mediated repression. Cell Death Differ 22:22–33
Suarez Y, Sessa WC (2009) MicroRNAs as novel regulators of angiogenesis. Circ Res 104:442–454
Urbich C, Kuehbacher A, Dimmeler S (2008) Role of microRNAs in vascular diseases, inflammation and angiogenesis. Cardiovasc Res 79:581
Khurana R, Simons M et al (2005) Role of angiogenesis in cardiovascular disease a critical appraisal. Circulation 112:1813–1824
Anastasiadou E, Jacob LS, Slack FJ (2018) Non-coding RNA networks in cancer. Nat Rev Cancer 18:5
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16:203–222
Wojciechowska A, Braniewska A, Kozar-Kamińska K (2017) MicroRNA in cardiovascular biology and disease. Adv Clin Exp Med 26:865–874
Tribolet L, Kerr E et al (2020) MicroRNA biomarkers for infectious diseases: from basic research to biosensing. Front Microbiol 11:1197
Hu S, Zhu W et al (2014) MicroRNA-155 broadly orchestrates inflammation-induced changes of microRNA expression in breast cancer. Cell Res 24:254–257
Eis PS, Tam W et al (2005) Accumulation of MiR-155 and BIC RNA in human b cell lymphomas. Proc Natl Acad Sci U S A 102:3627–3632
Takahashi RU, Miyazaki H, Ochiya T (2015) The roles of microRNAs in breast cancer. Cancers (Basel) 7:598–616
Nana-Sinkam SP, Croce CM (2011) MicroRNAs as therapeutic targets in cancer. Transl Res 157:216–225
Wang S, Xiao Y et al (2018) A gapmer aptamer nanobiosensor for real-time monitoring of transcription and translation in single cells. Biomaterials 156:56–64
Wang S, Majumder S et al (2018) Simultaneous monitoring of transcription and translation in mammalian cell-free expression in bulk and in cell-sized droplets. Synth Biol 3:ysy005
Wang S, Sun J et al (2016) A nanobiosensor for dynamic single cell analysis during microvascular self-organization. Nanoscale 8:16894–16901
Wang S, Riahi R et al (2015) Single cell nanobiosensors for dynamic gene expression profiling in native tissue microenvironments. Adv Mater 27:6034–6038
Jusiak B, Cleto S et al (2016) Engineering synthetic gene circuits in living cells with CRISPR technology. Trends Biotechnol 34:535–547
Ausländer S, Fussenegger M (2017) Synthetic RNA-based switches for mammalian gene expression control. Curr Opin Biotechnol 48:54–60
Matsuura S, Ono H et al (2018) Synthetic RNA-based logic computation in mammalian cells. Nat Commun 9:1–8
Rossetti M, Del Grosso E et al (2019) Programmable RNA-based systems for sensing and diagnostic applications. Anal Bioanal Chem 411:4293–4302
Deng R, Tang L et al (2014) Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cells. Angew Chem 126:2421–2425
Shen C-C, Hsu M-N et al (2019) Synthetic switch to minimize CRISPR off-target effects by self-restricting cas9 transcription and translation. Nucleic Acids Res 47:e13–e13
Endo K, Hayashi K, Saito H (2016) High-resolution identification and separation of living cell types by multiple microRNA-responsive synthetic mRNAs. Sci Rep 6:1–8
Miki K, Endo K et al (2015) Efficient detection and purification of cell populations using synthetic microRNA switches. Cell Stem Cell 16:699–711
Hirosawa M, Fujita Y et al (2017) Cell-type-specific genome editing with a microRNA-responsive CRISPR–Cas9 switch. Nucleic Acids Res 45:e118–e118
Green AA, Silver PA et al (2014) Toehold switches: De-novo-designed regulators of gene expression. Cell 159:925–939
Mousavi PS, Smith SJ et al (2020) A multiplexed, electrochemical interface for gene-circuit-based sensors. Nat Chem 12:48–55
Pardee K, Green AA et al (2016) Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell 165:1255–1266
Da Silva SJR, Silva CT, a. D. et al (2020) Clinical and laboratory diagnosis of sars-cov-2, the virus causing covid-19. ACS Infect Dis 6:2319–2336
Wang S, Emery NJ, Liu AP (2019) A novel synthetic toehold switch for microRNA detection in mammalian cells. ACS Synth Biol 8:1079–1088
Cui W, Meng W et al (2019) TGF-β-induced long non-coding RNA MiR155hg promotes the progression and EMT of laryngeal squamous cell carcinoma by regulating the MiR-155-5p/sox10 axis. Int J Oncol 54:2005–2018
Xie F, Ling L et al (2018) TGF-β signaling in cancer metastasis. Acta Biochim Biophys Sin 50:121–132
Adams JM, Cory S (2018) The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ 25:27
Li D-P, Fan J et al (2017) MiR-155 up-regulated by TGF-β promotes epithelial-mesenchymal transition, invasion and metastasis of human hepatocellular carcinoma cells in vitro. Am J Transl Res 9:2956
Louafi F, Martinez-Nunez RT, Sanchez-Elsner T (2010) MicroRNA-155 targets smad2 and modulates the response of macrophages to transforming growth factor-β. J Biol Chem 285:41328–41336
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Zhao, Y., Poudel, P., Wang, S. (2024). Detection of MicroRNAs Using Synthetic Toehold Switch in Mammalian Cells. In: Ceroni, F., Polizzi, K. (eds) Mammalian Synthetic Systems. Methods in Molecular Biology, vol 2774. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3718-0_16
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3718-0_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3717-3
Online ISBN: 978-1-0716-3718-0
eBook Packages: Springer Protocols