Abstract
Direct reprogramming of fibroblasts into induced cardiomyocytes (iCMs) through microRNAs (miRNAs) is a new emerging strategy for myocardial regeneration after ischemic heart disease. Previous studies have reported that murine fibroblasts can be directly reprogrammed into iCMs by transient transfection with four miRNAs (miRs-1, 133, 208 and 499 – termed “miRcombo”). While advancement in the knowledge of direct cell reprogramming molecular mechanism is in progress, it is important to investigate if this strategy may be translated to humans. Recently, we demonstrated that miRcombo transfection is able to induce direct reprogramming of adult human cardiac fibroblasts (AHCFs) into iCMs. Although additional studies are needed to achieve iCM maturation, our early findings pave the way toward new therapeutic strategies for cardiac regeneration in humans. This chapter describes methods for inducing direct reprogramming of AHCFs into iCMs through miRcombo transient transfection, showing experiments to perform for assessing iCM generation.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- microRNAs
- Transient transfection
- Direct cell reprogramming
- Human cardiac fibroblasts
- Induced cardiomyocytes
1 Introduction
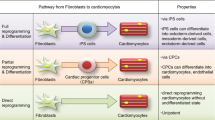
Myocardial infarction (MI) is the leading cause of death worldwide, causing massive loss of cardiomyocytes (CMs), inflammation and the formation of a non-functional fibrotic scar [1]. The poor regenerative capacity of adult cardiac tissue is the major barrier for heart regeneration. After MI, fibroblasts are the dominant cell type populating the cardiac scar [2]. Therefore, direct reprogramming of fibroblasts into iCMs provides a good therapeutic strategy to restore cardiac functions by repopulating cardiac scar with functional CMs [3]. In 2012, Jayawardena et al. have reported a combination of four miRNAs (miRcombo), able to induce direct reprogramming of mouse neonatal cardiac fibroblasts into iCMs [4]. Delivery of miRcombo was performed through cell transient transfection using the commercial transfection agent DharmaFECT1, and demonstrating that a single miRcombo dose may establish and maintain CM phenotype in cardiac fibroblasts [4]. Later, we have demonstrated that miRcombo is also able to directly reprogram AHCFs into iCMs , showing cardiomyocyte gene expression after 7 and 15 days of culture, 11% of reprogramming efficiency after 15 days of culture and spontaneous calcium transients after 30 days [5]. Here, we describe our protocol for inducing AHCF reprogramming into iCMs through single miRcombo transient transfection (Fig. 1).
Experimental outline for microRNA-mediated reprogramming of AHCFs into iCMs experiments including cell seeding and miRNA transient transfection using DharmaFECT1. We also indicated the expected timeline for induction of cardiomyocyte TFs (day 7), cardiomyocyte and fibroblast markers assessed by ddPCR expression or flow cytometry (day 15) and calcium transients analysis (day 30). (Figure created with Biorender.com under license)
2 Materials
2.1 Reagents for Cell Culture
-
1.
Adult human cardiac fibroblasts (AHCFs) cryopreserved (Lonza).
-
2.
Fibroblast expansion media: Fibroblast basal growth medium (FGM-3, Lonza), 10% fetal bovine serum (FBS), 0.1% insulin, 0.1% human fibroblast growth factor (hFGF), 0.1% gentamicin amphotericin (GA-1000).
-
3.
Reprogramming media: Dulbecco’s Modified Eagle Medium (DMEM) with 4.5 g/L glucose (without L-glutamine), 10% fetal bovine serum, 1% L-glutamine, 1% penicillin/streptomycin (when specified in the text).
-
4.
0.05% Trypsin/EDTA.
2.2 Reagents for microRNA Transfection
-
1.
DharmaFECT1 Transfection Reagent (Dharmacon).
-
2.
Synthetic mature miRNA mimics: hsa-miR-1a-3p, hsa-miR-133a-3p, hsa-miR-208a-3p, hsa-miR-499a-5p, mirVana™ miRNA Mimic Negative Control #1 (mirVana).
2.3 Reagents for RNA Purification and cDNA Synthesis
-
1.
QIAzol Lysis Reagent (QIAgen).
-
2.
Chloroform.
-
3.
70% ethanol in H2O stored at −20 ° C.
-
4.
100% 2-propanol stored at −20 ° C.
-
5.
DNase- and RNase-free H2O.
-
6.
High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem).
-
7.
miRCURY LNA RT Kit (Bio-Rad).
2.4 Reagents for Droplet Digital PCR
All reagents and instruments are from Bio-Rad:
-
1.
DG8 Cartridges QX200.
-
2.
DG8 Gaskets QX200.
-
3.
QX200 Droplet Generator.
-
4.
QX200 Droplet Reader.
-
5.
QX200 droplet generation oil for EvaGreen assays .
-
6.
Droplet generation oil for probes.
-
7.
QX200 ddPCR EvaGreen Supermix.
-
8.
ddPCR supermix for probes without dUTP.
-
9.
ddPCR Gene Expression Assay (human) primers (Table 1).
2.5 Reagents for Flow Cytometry
-
1.
0.05% Trypsin/EDTA.
-
2.
Phosphate buffer saline (PBS), 0.5% Tween-20 (v/v).
-
3.
PBS, 10% FBS, 1% sodium azide.
-
4.
Primary antibody for cardiac troponin T (Invitrogen).
-
5.
Fluorescent-labelled secondary antibody.
2.6 Reagents for Calcium Transient Imaging
-
1.
Modified Tyrode’s solution: 140 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 10 mM Hepes, in 2 mL of H2O, 0.1% bovine serum albumin (BSA).
-
2.
Fluo-4, cell permeant (Invitrogen) used at final concentration of 5 μM in Dimethyl Sulfoxide (DMSO).
-
3.
35 mm cell treated-bottom μ-dishes (Ibidi).
3 Methods
3.1 Culture of Adult Human Cardiac Fibroblasts
-
1.
Under laminar flow hood, for each cryovial containing the frozen cells, prepare 6 mL of Fibroblast expansion media in a sterile 15-mL conical tube and place it at 37 °C.
-
2.
Remove the cryovial from liquid nitrogen and place it in a 37 °C water bath. Transfer the cells into the tube containing the pre-warmed complete fibroblast expansion media from previous step.
-
3.
Centrifuge at 1000× g for 5 minutes at room temperature, then discard the supernatant and resuspend the cell pellet with fresh complete fibroblast expansion media Plate the cells in a 10-cm Petri dish and place in 37 °C incubator with humidified atmosphere of 5% CO2.
-
4.
When cells are 80% confluent, split them from passage 1 (P1) to P2 at a ratio of 1:3 by using 1 mL of 0.05% Trypsin/EDTA. Subsequently, split cells until P5. Change media every two days. Do not conduct microRNA transfections experiments in cells beyond P5.
3.2 AHCF Transfection with microRNAs
-
1.
Plate AHCFs in antibiotic-free reprogramming media. Transfections can be performed in 6-well (RNA), 12-well (flow cytometry) or 35-mm dish (calcium transient) format, depending on the nature of the experiment .
-
2.
Set up the transfection reaction under completely antibiotic-free medium conditions. Start by preparing two tubes: (tube 1) dilution of microRNAs 5 μM in serum-free DMEM and (tube 2) dilution of DharmaFECT1 in serum-free DMEM. Use the proportions outlined in the table below (Table 2). Incubate at room temperate for 5 minutes. Then, add tube 1 content into tube 2, mix twice and incubate at room temperature for 20 min (transfection mix). In order to compare and validate the relative induction of cardiac reprogramming, transfect cells in controls versus microRNA-treated samples.
-
3.
During the incubation time, change medium on cells to the appropriate volume of antibiotic-free reprogramming media as indicated in the table below (Table 2).
-
4.
Add transfection mix directly to cells.
-
5.
After 24 h, remove medium, rinse cells with PBS and add fresh complete reprogramming media. Continue cell culture up to desired time point (2, 7, 15 or 30 days post-transfection).
3.3 RNA Extraction
-
1.
Remove medium from cells without washing the cells.
-
2.
Under chemical hood, add 1 mL of QIAzol to each well and homogenize the cells. Collect cell lysate in 2-mL tube and either freeze lysate at −80 °C or proceed immediately to RNA purification. During all steps, take samples in ice bath.
-
3.
Add 200 μL of chloroform, vortex and centrifuge RNA at 12400× g at 4 °C for 30 min.
-
4.
Ensure efficient phase separation of samples and transfer the upper aqueous phase into a fresh 2-mL tube.
-
5.
Add 500 μL of ice-cold 2-propanol, invert tube twice and store at −20 °C overnight.
-
6.
Centrifuge at 12400× g for 30 minutes at 4 °C. The RNA precipitate will form a pellet on the side and bottom of the tube.
-
7.
Remove the supernatant and wash pellets twice with 1 mL of ice-cold 70% ethanol at 7400× g for 20 min at 4 °C.
-
8.
Remove supernatant and briefly dry the RNA pellet for 10 min by air-drying or under a vacuum.
-
9.
Add an appropriate volume of DNase and RNAse free H2O and store at −20 °C.
-
10.
Assess RNA concentration and quality by absorbance ratio at 260 nm/280 nm (A260/A280) and 260 nm/230 nm (A260/A230) wavelength. RNA concentration should be around 400 ng/μl with typical requirements of A260/A280 ratio >1.8–2.2 and A260/A230 ratio >1.7.
3.4 Evaluation of Transfection Efficiency: Day 2
-
1.
To study effective miRNA transfection, plate AHCFs in 6-multiwell in antibiotic-free reprogramming media.
-
2.
The day after, transfect cells with miR-1 and negmiR using DharmaFECT1 following Table 2 (see Note 1). Use untransfected AHCFs as controls.
-
3.
After 24 h from transfection, remove medium, wash cells with PBS and incubate cells with fresh complete reprogramming media.
-
4.
Perform RNA extraction as described in Subheading 3.3.
-
5.
Perform miRNA reverse-transcription using miRCURY LNA RT Kit, diluting RNA samples to 5 ng/μl in DNase and RNAse free H2O and preparing the reaction following the manufacturer’s instructions.
-
6.
Perform ddPCR analysis to assess the expression of miR-1 using EvaGreen supermix and primer for miR-1, following manufacturer’s instructions.
-
7.
Set thermal-cycling conditions: 95 °C for 5 minutes (1 cycle), 95 °C for 30 s and 55 °C for 1 minute (40 cycles), 90 °C for 5 minutes (1 cycle), and a 4 °C infinite hold. Load PCR plate on Bio-Rad QX200 droplet reader for quantification of cDNA copies/μl and analyze data using QuantaSoft analysis software.
-
8.
In parallel, prepare cDNA synthesis to assess TWF-1 (miR-1 target) knockdown using High capacity cDNA kit. Dilute RNA to 200 ng in DNase and RNAse free H2O and prepare the reaction following the manufacturer’s instructions.
-
9.
Perform ddPCR analysis using ddPCR supermix for probes (without dUTP) and primers for TWF-1. Use GAPDH primer as housekeeping gene.
-
10.
Set thermal-cycling conditions: 95 °C for 10 min (1 cycle), 94 °C for 30 s and 55 °C for 30 s (40 cycles), 98 °C for 10 minutes (1 cycle), and a 4 °C infinite hold. Load PCR plate on Bio-Rad QX200 droplet reader for quantification of cDNA copies/μL and analyze data using QuantaSoft analysis software.
-
11.
Expected miR-1 expression in miR-1 transfected cells should be around 500-fold compared to controls. Expected TWF-1 mRNA expression in miR-1 transfected AHCFs should be between 10% and 20% compared to negmiR controls and untransfected cells.
3.5 Evaluation of Cell Reprogramming: Day 7
-
1.
To study cardiomyocyte transcription factor (TF) expression in miRcombo-transfected cells, plate AHCFs in 6-multiwell in antibiotic-free reprogramming media. The day after, transfect cells with miRcombo and negmiR using DharmaFECT1 following Table 2. Use untransfected AHCFs as controls (see Note 3). After 24 h from transfection, remove medium, wash cells with PBS and incubate cells with fresh complete reprogramming media.
-
2.
After 7 days of culture, harvest RNA and synthesize total cDNA using High capacity cDNA. Dilute RNA to 200 ng in DNase and RNAse free H2O and preparing the reaction following the manufacturer’s instructions.
-
3.
For the first evidence of fibroblast reprogramming into iCMs in miRcombo-transfected cells, assess the expression of GATA4, MEF2C, TBX5, HAND2 and NKX2.5 cardiomyocyte TF expression performing ddPCR using ddPCR supermix for probes (without dUTP). Set thermal-cycling conditions as reported in Subheading 3.4, step 10. With the exception of NKX2.5 TF, whose upregulation is two-fold higher compared to controls, the expression of cardiomyocyte TFs in miRcombo-transfected cells is expected to be three- to six- fold higher compared to negmiR and untransfected control cells.
3.6 Evaluation of Cell Reprogramming: Day 15
-
1.
To study cardiomyocyte and fibroblast marker expression and reprogramming efficiency after 15 days of culture, plate cells for RNA extraction (6-multiwell) and flow cytometry (12-multiwell) in antibiotic-free reprogramming media. The day after, transfect cells with miRcombo and negmiR using DharmaFECT1 following Table 2. Use untransfected AHCFs as controls. After 24 h from transfection, remove medium, wash cells with PBS and incubate cells with fresh complete reprogramming media.
-
2.
After 15 days of culture, harvest RNA and synthesize total cDNA using High capacity cDNA. Dilute RNA to 200 ng in DNase and RNAse free H2O and preparing the reaction following the manufacturer’s instructions.
-
3.
For mature cardiomyocyte markers, assess the expression of TNNT2 and TNNI3 genes through ddPCR using ddPCR supermix for probes (without dUTP). Set thermal-cycling conditions as reported in Subheading 3.4, step 10. Cardiomyocyte gene expression in miRcombo transfected cells should reach two- to three-fold compared to negmiR and untransfected controls. For fibroblast-associated markers, assess the expression of VIM, DDR2 and FSP-1 genes. Expected fibroblast gene expression in miRcombo transfected cells should be between 30% and 60% compared to negmiR and untransfected control cells.
-
4.
In parallel, perform flow cytometry analysis to assess reprogramming efficiency in miRcombo and negmiR transfected cells.
-
5.
Remove medium and wash cells with PBS. Trypsinize cells with 500 μL of 0.05% Trypsin/EDTA and permeabilize cells with 0.5% v/v Tween 20 in PBS for 5 minutes.
-
6.
Wash cells with ice cold PBS with 10% FBS and 1% sodium azide.
-
7.
Incubate cells with Cardiac Troponin T primary antibody for 1 h at 4 °C. Then, wash samples twice in PBS with 10% FBS and 1% sodium azide and centrifuge at 1100× g for 5 min.
-
8.
Incubate samples with fluorescently-labelled secondary antibody (choose proper fluorophore according to laser excitation wavelength) for 1 h at 4 °C in the dark. Then, wash samples twice in PBS with 10% FBS and 1% sodium azide and centrifuge.
-
9.
Evaluate cell reprogramming using flow cytometry. Percentage of cTnT+ miRcombo transfected cells is expected between 9% and 12% compared to negmiR control cells, which do not show cTnT+ cells. Use control cells stained only with secondary fluorescent antibody to set background fluorescence.
3.7 Evaluation of Cell Reprogramming: Day 30
-
1.
To analyse calcium transient in miRcombo-transfected AHCFs, plate cells in 35 mm cell treated-bottom μ-dishes in antibiotic-free reprogramming media. The day after, transfect cells with miRcombo and negmiR using DharmaFECT1 and following Table 2. Use untransfected AHCFs as controls. After 24 h from transfection, remove medium, wash cells with PBS and incubate cells with fresh complete reprogramming media.
-
2.
After 30 days of culture, remove medium and wash cells twice with PBS. Load cells with 5 μM of Fluo-4 AM in modified Tyrode’s solution at 37 °C for 30 min while shielded from light.
-
3.
Wash cells twice in modified Tyrode’s solution and incubate them at 37 °C for 30 min to allow complete de-esterification of intracellular compounds.
-
4.
Record calcium transient using fluorescence microscope and performing high-speed time lapse lasting at least 2 min.
-
5.
Analyse calcium transients using ImageJ software (NIH) and report calcium level as F/F0 ratio, where F is the intensity of fluorescence emission recorded for each cell, while F0 is the background fluorescence.
4 Notes
-
1.
To rapidly assess cell transfection efficiency with DharmaFECT1, fluorescent-labelled microRNA mimic can be used. Cell transfection can be assessed 24 h post transfection using flow cytometry. Use cells incubated with fluorescent-labelled microRNA (without DharmaFECT1) and untransfected cells as negative controls.
-
2.
It is recommended to prepare 50 μM miRNA mimic stock solution in nuclease free H2O. This will reduce miRNA degradation during freeze-thaw cycles.
-
3.
In the case of miRcombo transfection (miR-1, miR-133a, miR-208a, miR-499a-5p), add ¼ of the final volume indicated in the Table 2 for each miRNAs.
Change history
14 January 2023
This chapter was previously published as non-Open Access. Under the request of the authors the chapter is now converted to Open Access and is available as Open Access under a CC BY 4.0 license.
References
Talman V, Ruskoaho H (2016) Cardiac fibrosis in myocardial infarction-from repair and remodeling to regeneration. Cell Tissue Res 365:563–581
Hinderer S, Schenke-Layland K (2019) Cardiac fibrosis – a short review of causes and therapeutic strategies. Adv Drug Deliv Rev 146:77–82
Sadahiro T, Yamanaka S, Ieda M (2015) Direct cardiac reprogramming. Circ Res 116:1378–1391
Jayawardena TM, Egemnazarov B, Finch EA et al (2012) MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 110:1465–1473
Paoletti C, Divieto C, Tarricone G et al (2020) MicroRNA-mediated direct reprogramming of human adult fibroblasts toward cardiac phenotype. Front Bioeng Biotechnol 8(529):1–14
Funding
The project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programe (Grant Agreement No. 772168).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this protocol
Cite this protocol
Paoletti, C., Divieto, C., Chiono, V. (2022). Direct Reprogramming of Adult Human Cardiac Fibroblasts into Induced Cardiomyocytes Using miRcombo. In: Ishikawa, K. (eds) Cardiac Gene Therapy. Methods in Molecular Biology, vol 2573. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2707-5_3
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2707-5_3
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2706-8
Online ISBN: 978-1-0716-2707-5
eBook Packages: Springer Protocols