Abstract
Scleractinians, or stony corals, are colonial animals that possess a high regenerative capacity and a highly diverse innate immune system. As such they present the opportunity to investigate the interconnection between regeneration and immunity in a colonial animal. Understanding the relationship between regeneration and immunity in stony corals is of further interest as it has major implications for coral reef health. One method for understanding the role of innate immunity in scleractinian regeneration is in situ hybridization using RNA probes. Here we describe a protocol for in situ hybridization in adult stony corals using a digoxigenin (DIG)-labeled RNA antisense probe which can be utilized to investigate the spatial expression of immune factors during regeneration.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Scleractinian corals are stony corals that build coral reefs. They are part of Cnidaria, a diverse phylum that possesses over >10,000 known species and is the sister group to Bilateria. It is estimated that these groups split approximately 604–748 million years ago (Fig. 1) [1]. Stony corals are primarily colonial consisting of many clonal polyps that are interconnected through a web of gastrovascular canals [2]. Stony corals are known to possess a high capacity for tissue regeneration which is hypothesized to be driven by stem cell differentiation and proliferation. However, the mechanisms of this regeneration are still not well understood (Fig. 2) [3]. They also possess a highly diversified innate immune system [4, 5]. From the available coral genomes, we understand that many corals possess a high diversity of immune factors which originated both from neo- and subfunctionalization events. This indicates that a complex interaction of immune factors and regenerative factors may be involved in whole body regeneration [6,7,8,9,10].
Cnidarian phylogenetic tree. Scleractinians, or stony corals, are part of the phylum Cnidaria. This phylum is a diverse primarily marine phylum and is important for evolutionary study due to its placement as the sister group to Bilateria. The split between Bilateria and Cnidaria is estimated to have occurred 604–748 million years ago
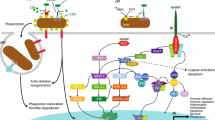
Schematic of whole-body regeneration in a colonial coral . During whole body regeneration of a colonial coral polyp, immune and stem cell factors are upregulated in response to the injury , and initiate regeneration of the body. Communication between the adjacent polyps and the regenerating polyp are critical for whole body regeneration. Stem and immune cells from adjacent polyps are presumed to migrate into the regenerating polyp area. Depending on the coral species and environmental conditions whole polyp regeneration can take 7–30+ days [3]
The underlying functional mechanisms of whole-body regeneration in stony corals is not as well understood as in other cnidarian models such as Nematostella and Hydra [3]. This is due to a historical emphasis within coral rsearch to focus on the heat stress response, challenges of manipulating the porous stony skeleton, and the high amounts of obligate microbial symbioses [11,12,13]. But with recent advances in sequencing technologies and cell biological techniques many of these challenges are starting to be addressed and tools to study whole-body regeneration are being developed [14,15,16]. Many genes and proteins have now been identified as involved in whole-body regeneration and immunity in corals, however the functional mechanisms of many of these genes are not known (Table 1) [3, 6].
One of the ways to investigate the interplay of innate immunity and regeneration is to use the method in situ hybridization (ISH) of RNA probes to assess the spatial gene expression of specific genes of interest. ISH is a very versatile technique because the RNA probes can be designed for any gene that is expressed. This method was first developed for the study of embryogenesis and has been further developed to understand the spatial gene expression across different live stages and stress response of many different organisms [17]. This method can be done by using many different types of probes labeled with nonradioactive nucleotides such as digoxigenin (DIG), fluorophores, or radioactive nucleotides [17,18,19]. The power of this technique is that it can detect and visualize small amounts of RNA at a cellular level. This is particularly useful for nonmodel organisms where the link between cell types and specific gene expression may not be understood.
To use this technique to study immunity during whole body regeneration, immune stimulation can be performed using synthetic elicitors such as lipopolysaccharides, peptidoglycans or exposure to known pathogens such as Vibrio to target immune and stem cell related genes that may be expressed during regeneration (Table 1) [20,21,22,23,24]. Additionally, no immune stimulator may be necessary if investigating the early process of regeneration, as the early signals of regeneration are early wound healing gene related to innate immunity [25].
In preparation for this method, DIG-labeled RNA antisense probe, and its accompanying sense control probe should already be designed and ready for use. Additionally, serial sections of paraffin embedded tissue should be prepared for testing both the sense and antisense probes. The sense probe is used as a control for nonspecific binding. If the sense probe has positive staining , then it will indicate that your antisense probe is not targeting the intended RNA. DIG-labeled probes are highly sensitive and can be developed from expressed RNAs for many different stressors [26,27,28,29,30]. It can also be applied to a wide range of tissues and organisms. This technique is not new, however, the application of it on adult stony coral tissues is an emerging technique that has promise for understanding the spatial expression of genes associated with whole-body regeneration. While this technique has primarily been used to assess the expression of developmental genes in cnidarians embryos and larvae, it has recently been modified to be used for adult cnidarians including stony corals [29]. In this book chapter we will outline the steps for performing in situ hybridization on stony coral tissue slices to yield cellular level resolution. This procedure could be easily modified for other cnidarians such as Nematostella and Aiptasia.
2 Materials
All solutions and dilutions should be made using molecular grade, RNase-free reagents, equipment and consumables. This procedure is highly sensitive to RNase contamination which can degrade the RNA probe.
2.1 Removal of Paraffin
-
1.
Thin-sectioned paraffin-embedded slides.
-
2.
100% xylene.
-
3.
Glass Coplin jars (see Note 1).
-
4.
100% ethanol.
-
5.
80% (v/v) ethanol.
-
6.
70% (v/v) ethanol.
-
7.
60% (v/v) ethanol.
-
8.
Sterile slide mailer.
-
9.
Hot water bath set to 100 °C.
-
10.
37 °C incubator.
-
11.
10× phosphate buffered saline (PBS): 2.56 g NaH2PO4, 11.94 g Na2HPO4, 102.2 g NaCl in 1 L. The phosphates should initially be mixed in 800 mL of water for a 1 L volume. pH should be 7.4 ± 0.4, if not, adjust pH to 7.4 with NaOH or HCl. After pH is adjusted, autoclave the solution. Once autoclaved, add NaCl and the last of the water to bring the final volume to 1 L.
-
12.
1× PBS: 100 mL 10× PBS, 900 mL water.
-
13.
10 mg/mL proteinase K stock solution: 10 mg proteinase K in 10 mL 1× PBS. Aliquot and store at −20 °C for later use.
-
14.
Proteinase K working solution: 90 μL proteinase K stock solution, 18 mL 1× PBS. Prepare fresh (see Note 2).
-
15.
20× saline sodium citrate (SSC): 175.3 g NaCl, 88.2 g Na3C6H5O7, 800 mL water. Adjust pH to 4.5 using HCl, bring to a final volume of 1.0 L and autoclave.
-
16.
50× Denhardt’s: 1% (w/v) Ficoll 400, 1% (w/v) polyvinylpyrrolidone (PVP), 1% (w/v) bovine serum albumin (BSA). Bring to a final volume of 100 mL using water. Dissolve all components in the water by stirring with a magnetic stirrer and filter any particulates. Store at −20 °C prior to use.
-
17.
20% (v/v) Tween 20: 20 mL Tween 20, 80 mL water. Mix Tween 20 in water using a stir plate. Store at 4 °C prior to use.
-
18.
20% (w/v) sodium dodecyl sulfate (SDS): 900 mL water, 200 g SDS. Bring this mixture to 68 °C and stir using a magnetic stirrer. Adjust the pH to 7.2 using HCl. Once the solution is completely mixed, adjust the volume to 1 L using water and store at room temperature.
-
19.
Prehybridization (prehybe) buffer: 20 mL formamide, 12.5 mL 20× SSC, 100 μL 20 mg/mL heparin, 5 mL 50× Denhardt’s, 20% (v/v) Tween 20, 20% (v/v) SDS, 2 mL of denatured salmon sperm DNA, 4.4 mL water. Aliquot in 50 mL tubes and keep at −20 °C for future use (see Note 3).
-
20.
Ice bath.
-
21.
0.2% (w/v) glycine–PBS solution: 1 g glycine, 45 mL 10× PBS, 405 mL water. Mix at room temperature using a stir plate to make sure that the glycine is fully dissolved.
-
22.
2× SSC solution: 20 mL 20× SSC, 180 mL water.
2.2 Hybridization of RNA Probe
-
1.
Hybridization buffer: 25 mL formamide, 12.5 mL 20× SSC, 100 μL 20 mg/mL heparin, 5 mL 50× Denhardt’s, 500 μL 20% (v/v) Tween 20, 500 μL 20% (w/v) SDS, 2 mL denatured salmon sperm DNA, 1 mL RNase-free water (see Note 3). Aliquot in 50 mL tubes and store at −20 °C for future use.
-
2.
RNA Probe (see Note 4).
-
3.
Hybridization-probe solution: 0.5 μL probe, 24.5 μL hybridization buffer. Prepare just prior to probe hybridization.
-
4.
Heat block set to 86–90 °C.
-
5.
PAP pen.
-
6.
Plastic coverslips.
-
7.
Slide moisture chamber.
-
8.
1× SSC solution: 10 mL 20× SSC, 190 mL water.
-
9.
0.5× SSC solution: 5 mL 20× SSC, 195 mL water.
-
10.
50% formamide: 250 mL formamide, 250 mL water.
-
11.
4× SSC + 50% formamide: 6 mL 20× SSC, 24 mL 50% formamide. Prepare fresh under a hood just prior to use.
2.3 Visualization of RNA Probe
-
1.
NaCl–Tris solution: 0.5 M NaCl, 0.5 M Tris. Bring total volume up to 50 mL in water.
-
2.
Alkaline phosphatase buffer (AP-buffer) without MgCl2: 20 mL NaCl–Tris solution, 500 μL Tween 20. Bring total volume up to 100 mL in water. Prepare just prior to use.
-
3.
AP-buffer: 50 mM MgCl2 in AP-buffer without MgCl2. Prepare just prior to use.
-
4.
DIG Nucleic Acid Detection Kit (e.g., Roche). Typically, all solutions should be prepared fresh just prior to use.
-
5.
AP Substrate Solution: 25 mL AP-Buffer, 82.5 μL nitro blue tetrazolium chloride (NBT), 82.5 μL 5-bromo-4-chloro-3-indolyl phosphate (BCIP). Made in the low light conditions and prepared just prior to use in a 50 mL tube. Cover with foil to keep in dark (see Note 5).
-
6.
Tris-EDTA (TE) buffer: 10 mM EDTA, pH 8.0, 1 mM Tris–HCl, pH 8.0, 98.8 mL water.
-
7.
Glycerol mounting medium: 80 μL 1× PBS, 20 μL glycerol.
3 Methods
All manipulations should be done using sterilized equipment and at room temperature, unless otherwise stated.
3.1 Removal of Paraffin
-
1.
Under a well-ventilated fume hood, pour 50 mL of 100% xylene into a sterile glass Coplin jar.
-
2.
Place tissue slides into the Coplin jar carefully.
-
3.
Allow 10 min for dewaxing to occur.
-
4.
Fill four sterile glass Coplin jars with 100% ethanol, 80% (v/v) ethanol, 70% (v/v) ethanol, and 60% (v/v) ethanol respectively.
-
5.
Soak slides in 100% ethanol for 10 min.
-
6.
Replace the 100% ethanol with fresh 100% ethanol.
-
7.
Incubate for 10 min.
-
8.
Move the slides, using sterile tweezers to the Coplin jar with 80% (v/v) ethanol.
-
9.
Incubate for 1 min.
-
10.
Move the slides, using sterile tweezers to the Coplin jar with 70% (v/v) ethanol.
-
11.
Incubate for 1 min.
-
12.
Move the slides, using sterile tweezers to the Coplin jar with 60% (v/v) ethanol.
-
13.
Incubate for 1 min.
3.2 Slide Pretreatment and Prehybe Preparation
-
1.
Set your incubator to 37 °C and turn on the hot water bath to 100 °C.
-
2.
Prepare the prehybe buffer by placing it in a boiling water bath for 15 min.
-
3.
Place the prehybe buffer in an ice bath for 5 min.
-
4.
Turn on your hybridization oven to the hybridization temperature (see Note 6).
-
5.
Add 18 mL of prehybe buffer to a new sterile slide mailer.
-
6.
Warm the slide mailer in the hybridization oven.
-
7.
Remove slides from the 60% (v/v) ethanol incubation using sterile tweezers.
-
8.
Place slides in a sterile slide mailer filled with 18 mL of 1× PBS.
-
9.
Wash for 5 min on an orbital shaker set to 100–150 rpm.
-
10.
Replace 1× PBS with 10 mL of proteinase K working solution to the slide mailer.
-
11.
Incubate without shaking for 15 min at 37 °C (see Note 7).
-
12.
Replace the proteinase K working solution in the slide mailer with 18 mL of 2× SSC solution.
-
13.
Incubate at 100–150 rpm for 10 min at room temperature.
-
14.
Transfer the slides to the slide mailer filled with the warmed prehybe buffer.
-
15.
Incubate the slides with the prehybe buffer at hybridization temperature for 1 h.
-
16.
Set the heat block to 86–90 °C.
3.3 RNA Probe Hybridization
-
1.
Prepare the hybridization-probe solution.
-
2.
Heat the hybridization-probe solution at 86–90 °C for 12 min using a heat block.
-
3.
Cool hybridization-probe solution for 1 min on ice.
-
4.
Remove the slide mailer from the hybridization oven.
-
5.
Carefully remove slides one by one using sterile tweezers, laying them flat on a paper towel.
-
6.
Remove excess prehybe buffer around samples.
-
7.
Use a PAP pen to encircle the tissue.
-
8.
Gently cover the tissue within the circle with 25 μL of hybridization-probe solution.
-
9.
Cover with plastic coverslip (see Note 8).
-
10.
Fill the bottom of the slide moisture chamber with 4× SSC + 50% formamide solution.
-
11.
Place slides on the slide moisture chamber.
-
12.
Place the slide moisture chamber in the hybridization oven set to 50–60 °C, depending on the hybridization of temperature of the probe (see Note 6).
-
13.
Incubate for 24 h.
-
14.
Carefully remove the slide moisture chamber from the hybridization oven.
-
15.
Open the slide moisture chamber, and carefully remove the coverslips from each slide using sterile tweezers, taking care to not displace the tissue (see Note 9).
-
16.
Rinse the slide with 1000 μL of 2× SSC solution by gently pipetting the solution to the tissue area using a pipette. Thoroughly rinse to ensure that all excess probes have been washed away to reduce nonspecific binding (see Note 10).
-
17.
Fill a sterile slide mailer with 18 mL of 2× SSC solution.
-
18.
Place each washed slide in the slide mailer.
-
19.
Incubate at room temperature for 5 min with gentle rotation on an orbital shaker.
-
20.
Replace the 2× SSC solution with 18 mL of 1× SSC solution.
-
21.
Incubate at room temperature for 5 min with gentle rotation on an orbital shaker.
-
22.
Set the incubator to 42 °C.
-
23.
Replace 1× SSC solution with 18 mL of fresh 1× SSC solution.
-
24.
Incubate at room temperature for 5 min with gentle rotation on the orbital shaker.
-
25.
Replace 1× SSC solution with 18 mL of 0.5× SSC.
-
26.
Incubate at 42 °C for 10 min without shaking.
-
27.
Replace 0.5× SSC solution with 18 mL of fresh 0.5× SSC solution.
-
28.
Incubate at 42 °C for 10 min without shaking.
3.4 RNA Probe Visualization
-
1.
Incubate slides for 1 min in 18 mL of AP-buffer without MgCl2 at room temperature.
-
2.
Block the slides overnight at 4 °C on an orbital shaker according to the DIG Nucleic Acid Detection Kit manufacturer’s instructions (see Note 11).
-
3.
Conjugate DIG for 3 h at room temperature with gentle shaking in a sterile slide mailer according to the kit’s instructions.
-
4.
Replace the conjugate buffer with 18 mL of AP-buffer without MgCl2.
-
5.
Incubate for 5 min at room temperature with gentle shaking.
-
6.
Replace the AP-buffer without MgCl2 with 18 mL of AP-buffer.
-
7.
Wash for 5 min at room temperature with gentle rotating on an orbital shaker.
-
8.
Replace AP-buffer with 18 mL of fresh AP-buffer.
-
9.
Wash for 5 min at room temperature with gentle rotating on an orbital shaker.
-
10.
Transfer slide mailer to a dark room.
-
11.
Replace the AP-buffer with 18 mL AP Substrate Solution.
-
12.
Incubate in the dark at room temperature.
-
13.
Check for purple color development every half hour (see Note 12).
-
14.
Once color development is present, transfer slides to a new sterile slide mailer with 18 mL TE buffer to stop color development .
-
15.
Incubate at room temperature in the dark for 5 min.
-
16.
Replace the TE buffer with 18 mL of RNase-free water.
-
17.
Incubate for 1 min at room temperature in the dark.
-
18.
Remove slides from the water, drying around the edges of the tissue.
-
19.
Add glycerol mounting medium and cover with coverslip.
-
20.
Store slides at 4 °C in the dark until images are taken (Fig. 3).
Representative example of in situ hybridization results in the pacific stony coral , Acropora hyacinthus to demonstrate general outcomes of specific and nonspecific binding. (a) This panel shows the staining of Chordin, a marker expressed during cnidarian regeneration [20] in Acropora hyacinthus tissue which has been exposed to a heat stress. The expression the antisense (−) probe for Chordin was found throughout the gastrodermis, and in gastrodermal cells. The sense (+) control probe showed some staining within the cnidocytes indicating nonspecific binding of cnidocytes. Cnidocytes are indicated by black arrows. (b) This panel shows the staining of Fructose Bisphosphate Aldolase again in Acropora hyacinthus tissue exposed to heat stress. The antisense (−) probe had primary staining within the cnidocyte cells in the epidermis. Cnidocytes are indicated by black arrows. The sense (+) control had no nonspecific staining indicating that the cnidocyte staining in the antisense (−) probe was specific
4 Notes
-
1.
Use glass jars for this step because xylene will melt plastic. Sterilize glass Coplin jar by autoclaving prior to use.
-
2.
Diluted proteinase K can degrade quickly so use a fresh aliquot each time.
-
3.
The salmon sperm needs to be denatured prior to adding to the prehybe buffer. To do this, boil an aliquot of the salmon sperm on a heat block for 5 min.
-
4.
RNA probes, both sense and antisense should be made prior to starting this protocol [29].
-
5.
As a substitute, BM Purple (Sigma-Aldrich, Germany) can be used. Keep in low light conditions.
-
6.
Hybridization temperatures vary greatly due to the probe that is being used. Most hybridization temperatures range from 50–60 °C. During this step make sure to work quickly so that you do not dry out the sample. Also be extremely careful to not touch the tissue sample. The tissue is still very delicate, and touching it, can disrupt the tissue integrity. Folding a paper towel or a Kimwipes, gently pat the area around the tissue to soak up the excess prehybe buffer.
-
7.
Do not shake the tissue samples at this step, as it can lead to damage of the tissue integrity.
-
8.
For best results use plastic coverslips. They are more flexible and easier to manipulate then glass coverslips.
-
9.
Each slide should be done one at a time, while the other slides are kept in the moisture chamber in the hybridization oven. Keeping the slides at room temperature with the probe still present will allow for more nonspecific binding.
-
10.
This step can be done over a sink or over a container to catch the 2× SSC solution. Gently pipette the solution over the slide, and let it rinse the slide. Make sure to rinse thoroughly, as the residual presence of a probe can lead to more nonspecific binding.
-
11.
A minimum of 1 h at room temperature with gentle shaking can also be done if constrained by time.
-
12.
In some cases, probe visualization may take longer than 24 h. If this the case, develop the sample at 4 °C checking every 12–24 h to see if probe development has progressed.
References
Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR (2006) The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol 7:R64. https://doi.org/10.1186/gb-2006-7-7-r64
Gladfelter EH (1983) Circulation of fluids in the gastrovascular system of the reef coral Acropora cervicornis. Biol Bull 165:619–636. https://doi.org/10.2307/1541469
Henry LA, Hart M (2005) Regeneration from injury and resource allocation in sponges and corals—a review. Int Rev Hydrobiol 90:125–158. https://doi.org/10.1002/iroh.200410759
Palmer CV, Traylor-Knowles NG (2018) Cnidaria: anthozoans in the hot seat. In: Advances in comparative immunology. Springer, Cham, pp 51–93
Palmer CV, Traylor-Knowles N (2012) Towards an integrated network of coral immune mechanisms. Proc R Soc B Biol Sci 279:4106–4114. https://doi.org/10.1098/rspb.2012.1477
Horricks RA, Herbinger CM, Lillie BN, Taylor P, Lumsden JS (2019) Differential protein abundance during the first month of regeneration of the Caribbean star coral Montastraea cavernosa. Coral Reefs 38:45–61. https://doi.org/10.1007/s00338-020-01952-4
Cunning R, Bay RA, Gillette P, Baker AC, Traylor-Knowles N (2018) Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Sci Rep 8:16134. https://doi.org/10.1038/s41598-018-34459-8
Shinzato C, Shoguchi E, Kawashima T et al (2011) Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476:320. https://doi.org/10.1038/nature10249
Voolstra CR, Li Y, Liew J et al (2017) Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci Rep 7:17583. https://doi.org/10.1038/s41598-017-17484-x
Shumaker A, Putnam HM, Qiu H et al (2019) Genome analysis of the rice coral Montipora capitata. Sci Rep 9:2571. https://doi.org/10.1038/s41598-019-39274-3
Blackall LL, Wilson B, van Oppen MJH (2015) Coral—the world’s most diverse symbiotic ecosystem. Mol Ecol 24:5330–5347. https://doi.org/10.1111/mec.13400
Cziesielski MJ, Schmidt-Roach S, Aranda M (2019) The past, present, and future of coral heat stress studies. Ecol Evol 9:10055–10066. https://doi.org/10.1002/ece3.5576
Parkinson JE, Baker AC, Baums IB et al (2020) Molecular tools for coral reef restoration: beyond biomarker discovery. Conserv Lett 13:e12687. https://doi.org/10.1111/conl.12687
Rosental B, Kozhekbaeva Z, Fernhoff N et al (2017) Coral cell separation and isolation by fluorescence-activated cell sorting (FACS). BMC Cell Biol 18:30. https://doi.org/10.1186/s12860-017-0146-8
Nowotny JD, Connelly MT, Traylor-Knowles N (2021) Novel methods to establish whole-body primary cell cultures for the cnidarians Nematostella vectensis and Pocillopora damicornis. Sci Rep 11:4086. https://doi.org/10.1038/s41598-021-83549-7
Levy S, Elek A, Grau-Bové X, Menéndez-Bravo S, Iglesias M, Tanay A, Mass T, Sebé-Pedrós A (2021) A stony coral cell atlas illuminates the molecular and cellular basis of coral symbiosis, calcification, and immunity. Cell 184:2973–2987.e18. https://doi.org/10.1016/j.cell.2021.04.005
Lehmann R, Tautz D (1994) Chapter 30. In situ hybridization to RNA. In: Methods in cell biology. Academic Press, New York, pp 575–598. https://doi.org/10.1016/S0091-679X(08)60933-4
Jin L, Lloyd RV (1997) In situ hybridization: methods and applications. J Clin Lab Anal 11:2–9. https://doi.org/10.1002/(SICI)1098-2825(1997)11:1
Levsky JM, Singer RH (2003) Fluorescence in situ hybridization: past, present and future. J Cell Sci 116:2833–2838. https://doi.org/10.1242/jcs.00633
Connelly MT, McRae CJ, Liu P, Traylor-Knowles N (2020) Lipopolysaccharide treatment stimulates Pocillopora coral genotype-specific immune responses but does not alter coral-associated bacteria communities. Dev Comp Immunol 109:103717. https://doi.org/10.1016/j.dci.2020.103717
Kvennefors ECE, Leggat W, Kerr CC, Ainsworth TD, Hoegh-Guldberg O, Barnes AC (2010) Analysis of evolutionarily conserved innate immune components in coral links immunity and symbiosis. Dev Comp Immunol 34:1219–1229. https://doi.org/10.1016/j.dci.2010.06.016
Vidal-Dupiol J, Ladrière O, Meistertzheim AL, Fouré L, Adjeroud M, Mitt AG (2011) Physiological responses of the scleractinian coral Pocillopora damicornis to bacterial stress from Vibrio coralliilyticus. J Exp Biol 214:1533–1545. https://doi.org/10.1242/jeb.053165
Zhi Z, Shuimiao Z, Jia T, Zhaoqun L, Yibo W, Yan W, Senjie L (2019) Altered immune landscape and disrupted coral-symbiodinium symbiosis in the scleractinian coral Pocillopora damicornis by vibrio coralliilyticus challenge. Front Physiol 10:366. https://doi.org/10.3389/fphys.2019.00366
Takagi T, Yoshioka Y, Zayasu Y et al (2020) Transcriptome analyses of immune system behaviors in primary polyp of coral Acropora digitifera exposed to the bacterial pathogen vibrio coralliilyticus under thermal loading. Marine Biotechnol 22:748–759. https://doi.org/10.1007/s10126-020-09984-1
Gurtner G, Werner S, Barrandon Y et al (2008) Wound repair and regeneration. Nature 453:314–321. https://doi.org/10.1038/nature07039
Darby IA, Bisucci T, Desmouliere A, Hewitson TD (2006) In situ hybridization using cRNA probes. In: Darby IA, Hewitson TD (eds) In situ hybridization protocols. Humana Press, Totowa, NJ. https://doi.org/10.1385/1-59745-007-3:17
Valentino KL, Eberwine JH, Barchas JD (1987) In situ hybridization. Oxford University Press, New York
Tautz D, Pfeifle C (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81–85. https://doi.org/10.1007/BF00291041
Traylor-Knowles N, Rose NH, Palumbi SR (2017) The cell specificity of gene expression in the response to heat stress in corals. J Exp Biol 220:1837–1845. https://doi.org/10.1242/jeb.155275
Wolenski FS, Layden MJ, Martindale MQ, Gilmore TD, Finnerty JR (2013) Characterizing the spatiotemporal expression of RNAs and proteins in the starlet sea anemone, Nematostella vectensis. Nat Protoc 8:900. https://doi.org/10.1038/nprot.2013.014
Palmer CV, Traylor-Knowles NG, Willis BL, Bythell JC (2011) Corals use similar immune cells and wound-healing processes as those of higher organisms. PLoS One 6:e23992. https://doi.org/10.1371/journal.pone.0023992
Acknowledgments
This work is supported by startup funds provided by University of Miami, Rosenstiel School of Marine and Atmospheric Sciences and by NSF-1951826. The authors would like to thank Traylor-Knowles Lab, as well as thank Bradford Dimos and Emily Buckley for discussions concerning the chapter development .
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this protocol
Cite this protocol
Traylor-Knowles, N., Emery, M. (2022). Analysis of Spatial Gene Expression at the Cellular Level in Stony Corals. In: Blanchoud, S., Galliot, B. (eds) Whole-Body Regeneration. Methods in Molecular Biology, vol 2450. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2172-1_19
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2172-1_19
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2171-4
Online ISBN: 978-1-0716-2172-1
eBook Packages: Springer Protocols