Abstract
The study of antigen receptor gene repertoires using next-generation sequencing (NGS) technologies has disclosed an unprecedented depth of complexity, requiring novel computational and analytical solutions. Several bioinformatics workflows have been developed to this end, including the T-cell receptor/immunoglobulin profiler (TRIP), a web application implemented in R shiny, specifically designed for the purposes of comprehensive repertoire analysis, which is the focus of this chapter. TRIP has the potential to perform robust immunoprofiling analysis through the extraction and processing of the IMGT/HighV-Quest output, via a series of functions, ensuring the analysis of high-quality, biologically relevant data through a multilevel process of data filtering. Subsequently, it provides in-depth analysis of antigen receptor gene rearrangements, including (a) clonality assessment; (b) extraction of variable (V), diversity (D), and joining (J) gene repertoires; (c) CDR3 characterization at both the nucleotide and amino acid level; and (d) somatic hypermutation analysis, in the case of immunoglobulin gene rearrangements. Relevant to mention, TRIP enables a high level of customization through the integration of various options in key aspects of the analysis, such as clonotype definition and computation, hence allowing for flexibility without compromising on accuracy.
Chrysi Galigalidou and Laura Zaragoza-Infante are equal first authors.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Antigen receptor
- B-cell receptor
- Immunoglobulin
- T-cell receptor
- Immunoinformatics
- Clonality
- Immune repertoire
- Somatic hypermutation
1 Introduction
Profiling the B-cell receptor immunoglobulin (BcR IG) and T-cell receptor (TR ) gene repertoires using next-generation sequencing (NGS) technologies advanced our understanding of various clinical conditions and biological processes, extending from infections, vaccination, autoimmunity, to malignancy. NGS immunogenetics has applications in both diagnostics (e.g., assessment of clonality in samples investigated for a possible lymphoproliferation or detection of minimal residual disease in patients with lymphoid malignancies) and research [1, 2]. To date, several pipelines that perform both BcR IG/TR sequence annotation and meta-data analysis have been made publicly available [3,4,5,6]: in that regard, notable examples include the “IMGT/StatClonotype” tool [7, 8], the MiXCR software [9], the Vidjil platform [10], and the ARReST|Interrogate application [11], among others.
Our contribution in this field concerns the T-cell receptor/immunoglobulin profiler (TRIP) software [12], which was designed in order to enable the comprehensive characterization of BcR IG and TR gene repertoires based on an integrated, robust, and user-friendly interface. TRIP has been utilized in projects on hematological malignancies, such as chronic lymphocytic leukemia (CLL) and multiple myeloma (MM) [13,14,15,16], as well as other contexts, e.g., infections [17, 18], providing valuable insight into the selection forces that shape the architecture of the respective immune repertoires.
This chapter will focus on the features of TRIP, particularly aiming to highlight how the functionalities offered by this software address the challenges of repertoire analysis in both diagnostic and, particularly, research settings.
2 Data Processing
2.1 Preprocessing of the Raw Data
The raw sequencing data is transferred from the sequencer server to the dedicated workspace.
The initial processing actions depend on the selected sequencing strategy; e.g., there might be need for some steps, such as demultiplexing, adapter masking, and format conversion to the FASTQ data type.
2.1.1 Demultiplexing
Demultiplexing concerns the separation of sequencing reads to the respective samples in cases of multiplexed sequencing, i.e., the simultaneous sequencing of multiple samples in a single run. During sample preparation, unique index sequences are attached to the sequences of each individual sample that will be used as identifiers during the demultiplexing process.
2.1.2 Adaptor Masking
Adapter sequence masking leads to the identification of adapter sequences and removes them from consideration in the downstream analysis steps. This process is essential in order to avoid artificial mismatches and alignment issues during sequence annotation.
2.1.3 Format Conversion
Relevant to mention, the output file(s) of this process are of the FASTQ type, since this is the format required for the downstream steps. The FASTQ file format is very commonly used in bioinformatics in order to process raw sequencing data, since it contains information regarding the sequence reads and their quality. FASTQ files contain four lines of information for each individual read:
-
1.
The first line begins with the “@” symbol followed by a read identifier, which is given during the sequencing process.
-
2.
The second line contains the nucleotide sequence of the read.
-
3.
The third line has a “+” symbol, used as a line separator.
-
4.
The fourth line has information about the quality of each base of the sequence, represented as Phred quality score. The value of these quality scores can be retrieved from ASCII charts.
Additional information about FASTQ files is provided at the following link https://www.ebi.ac.uk/ega/submission/sequence#fastq_format.
In the case of sequencing on an Illumina platform, the bcl2fastq2 software is the one most commonly used for demultiplexing sequencing data and for the masking of the adaptor sequences and/or UMIs (unique molecular identifiers), if present. Moreover, bcl2fastq2 transforms base call files (BCL), which is the default format of raw data when obtained from the Illumina sequencer platform, into FASTQ files. Some sequencing platforms, such as the MiniSeq or MiSeq, provide the option to automatically transform BCL files to the FASTQ format.
In case another sequencing platform is used, it is necessary to follow the instructions specified for each scenario, check the data format of the sequencer server output files, and transform them to FASTQ.
2.2 Filtering of the Raw Data
As a first step, quality filters should be applied to all reads in the FASTQ file(s) in order to ensure that only high-quality data will be subjected to further analysis. A set of filtering parameters can be selected according to the type of data and the design of the experiment. The reads that do not fulfill all the requirements will be filtered out. The most common parameters are related to the read length, the quality score of each individual nucleotide, and the overall quality score of each read.
The level of strictness of the parameters is chosen according to the overall quality of the NGS run and the minimum quality threshold that would allow the extraction of biologically meaningful results depending on the project design.
Indicative examples of parameters for the analysis of BcR IG /TR data include: minimum length of the raw reads, 150 nucleotides; quality threshold for each nucleotide, 14; accepted minimum mean sequence quality for each read, 20; maximum percentage of low-quality nucleotides, 0.2 (20%); and minimum percentage of accepted unidentified nucleotides, 0.01 (1%).
2.3 Synthesis of Paired-End Reads
Given the extreme intrinsic variability of BcR IG /TR rearrangement sequences, paired-end sequencing protocols are usually applied. In this scenario, two individual reads, namely, R1 and R2, are obtained from each sequence ensuring the high quality of the sequences and the accuracy of the immunogenetic annotation.
-
1.
After checking the quality of each individual read (see Subheading 2.2), perform the synthesis of full-length reads by merging the individual R1 and R2 reads corresponding to each sequence, through the identification of an overlapping region.
-
2.
Apply quality filters to the synthesized, full-length reads. Examples of these filters are: minimum length of the overlap between R1 and R2 reads, 20 nucleotides; mismatch ratio of the overlapping area, 0.25 (25%); threshold for the continuous match of the overlapping area, 10 nucleotides; quality thresholds for the classification of individual nucleotides either as of “bad quality” (and be replaced by “N”) or of “high quality”, 14 and 35, respectively; quality mean score of the synthesized reads, 25; minimum length of the synthesized reads, 280 nucleotides; percentage of nucleotides that can have low quality in the synthesized reads, 0.15 (15%); quality threshold of individual nucleotides in the synthesized reads, 20; percentage of “bad quality” nucleotides in the synthesized reads excluding the CDR3 , 0.005 (0.5%); and CDR3 quality threshold, 25. This set of filtering criteria was designed in order to take into account the intrinsic properties of the BcR IG /TR rearrangement sequences, i.e., the extreme variability of the CDR3 .
-
3.
Compare the number of synthesized reads that have passed all filters with the number of raw reads, and check the percentage of reads that is discarded due to each filter. If the percentage of the synthesized, high quality reads is low (e.g., below 50–60%), a revision of relevant filter(s) should be considered.
The final synthesized reads that have successfully passed all filters from each sample are deposited in a FASTA file. This file consists of two lines of information per sequence: the first line begins with a “>” symbol followed by the read identifier, and the second line contains the nucleotide sequence.
3 Sequence Annotation with IMGT/HighV-QUEST
IMGT (the international ImMunoGeneTics information system) is the worldwide reference in immunogenetics and immunoinformatics [19]. IMGT has incorporated the most extensive and updated reference datasets for human BcR IG /TR genes. IMGT/HighV-QUEST is the web portal for BcR IG /TR data analysis from NGS high-throughput and deep sequencing [20].
In the IMGT/HighV-QUEST home page (http://www.imgt.org/HighV-QUEST/home.action), the user can customize the analysis through a series of options, including a job title, the species, the antigen receptor type (BcR IG or TR ), and the specific locus (for instance, BcR IGH or IGL ). The data has to be uploaded in FASTA format, and the submission limit is 1,000,000 sequences. Once the analysis is finished, the results can be downloaded from the “Analysis history” tab.
The output for each sample is a folder with ten files in text (.txt) format, with each of them containing different types of immunogenetic information. More specifically, the output files are the following: “1_Summary.txt,” containing a summary table of basic immunogenetic information, such as the rearranged V(D)J genes, the % of identity with the germline, the presence of indels etc.; “2_IMGT-gapped-nt-sequences.txt”; “3_Nt-sequences.txt”; 4_IMGT _gapped_AA_sequences.txt”; “5_AA-sequences.txt; “6_Junction.txt”; “7_V-REGION-mutation-and-AA-change-table.txt”; “8_V-REGION-nt-mutation-statistics.txt”; “9_V-REGION-AA-change-statistics.txt”; “10_V-REGION-mutation-hotspots.txt”; “11_Parameters”, with the set of parameters applied in the analysis; and “README.txt”, with technical information about the analysis.
4 IMGT/HighV-QUEST Meta-Data Analysis with TRIP
The T-cell receptor/immunoglobulin profiler (TRIP) tool [12] is a web application that provides an in-depth meta-data analysis based on the processing of the IMGT/HighV-QUEST output files, through a number of interoperable modules. The TRIP tool can be downloaded from the following link: https://bio.tools/TRIP_-_T-cell_Receptor_Immunoglobulin_Profiler.
-
1.
Since IMGT/HighV-QUEST has a submission threshold of 1,000,000 sequences, if a sample contains a larger number of sequences, the user must split them into different batches of sequences before analyzing them with IMGT/HighV-QUEST. Thus, multiple output folders will be generated by the tool for the same sample. In this case, the folders should be named using the same identifier with a different extension, following a numerical order starting from 0, i.e., “-0”, “-1”, “-2”, etc. With this approach, TRIP can trace the origin of these files to the same sample and will combine the respective data for the analysis.
-
2.
The first step of the analysis with TRIP concerns the selection of the directory containing the IMGT/HighV-QUEST output data. At this step, it is also possible to restore previous sessions.
-
3.
The next step concerns data selection. TRIP allows for the simultaneous analysis of several datasets. In that case, the analysis is performed both individually for each dataset and for all datasets together (“All Data” output files). Moreover, if more than one dataset is selected, there will be additional available steps in the downstream analysis (such as the shared clonotype computation or the repertoire comparison, see below).
-
4.
The relevant output files from IMGT/HighV-QUEST are selected, depending on the type of downstream analysis. For most types of analysis, the necessary files are “1_Summary.txt,” “2_IMGT-gapped-nt-sequences.txt,” “4_IMGT-gapped-AA-sequences.txt,” and “6_Junction.txt.”
After loading the files (option “Load Data”), TRIP scans the data and gives a notification in the case of data headers with a different or an unknown value. In that case, data headers should be replaced with the appropriate ones.
-
5.
The antigen receptor type is selected: BcR IG or TR .
-
6.
The type of data to be analyzed is selected: high throughput (NGS data) or low throughout (Sanger sequencing data). Henceforth, this chapter will focus on the analysis of high-throughput data.
A summary of all aforementioned analytical steps is depicted in Fig. 1.
5 High-Throughput Data Analysis with TRIP
5.1 Preselection (Data Curation)
All the preselection filters should be applied:
-
1.
The Only take into account functional V-gene filter ensures the exclusive analysis of sequences utilizing a functional V gene; sequences with pseudogenes (P) or open reading frame (ORF) genes will be excluded from downstream analysis.
-
2.
The Only take into account CDR3 with no special characters filter removes from the analytical process sequences with characters others than those of the 20 amino acids.
-
3.
The Only take into account productive sequences filter limits the analysis to productive BcR IG /TR rearrangement sequences; sequences with stop codons or frameshifts will be filtered out.
-
4.
The filter entitled Only take into account CDR3 with valid start/end landmarks ensures that only sequences with well-annotated CDR3 will be subjected to further analysis. The CDR3 is delimited by a cysteine at IMGT position 104 (second-CYS 104) and a tryptophan or a phenylalanine (for BcR IG and TR sequences, respectively) at IMGT position 118 (i.e., J-PHE or J-TRP 118). If necessary, it is possible to add more than one landmark in the analysis by separating them with the “|” symbol.
Filters are applied consecutively and, as soon as one of the criteria is not passed, the sequence is filtered out; it is important to keep in mind that only the first non-passed criterion is reported. Only sequences that were of high quality according to all aforementioned standards will be further analyzed.
The results of the preselection process are summarized in four different tables and can be found at the “Pre-selection” tab:
-
1.
A summary of the included and excluded sequences based on the application of each criterion (“Summary”).
-
2.
The entire set of data (“All data table”).
-
3.
The set of filtered-in sequences that will be analysed in the following steps (“Clean table”).
-
4.
The set of filtered-out sequences (“Clean out table”).
The last data column in the “Clean out” table indicates the criterion that was not passed for each individual sequence. All tables can be downloaded in text (.txt) format.
5.2 Selection (Filtering)
Sequences, meeting all the pre-selection criteria, are further filtered during the Selection step.
The range of the V-region identity % should be selected. Sequences with a V region identity % that does not fall into the selected range are excluded from the analysis. The selection % of identity depends largely on:
-
1.
The type of antigen receptor gene sequence data, e.g., the SHM mechanism is operational exclusively in B cells.
-
2.
The expected error rate induced by the amplification protocol or the sequencing process.
In more detail, in the case of BcR IG sequences, a typical range of the V-region identity would be 85–100%, whereas in TR , the range would be narrower (95–100%).
The rest of the available filters enable the selection of sequences with specific immunogenetic features, namely, V, D, and J genes, CDR3 length and the presence of particular CDR3 amino acid sequence motifs. These filters allow for a high level of customization of the analytical procedure.
Again, four output files are produced, which are located at the “Selection” tab:
-
1.
A summary table with all filtered-in and -out sequences for each individual parameter (“Summary).
-
2.
The entire set of sequences that passed all the preselection criteria (“All Data table”).
-
3.
All sequences that passed through the Selection filters (“Filter in table”).
-
4.
The sequences that did not meet the selection criteria and were, thus, excluded from further analysis (“Filter out table”).
The last column of the “Filter out” table indicates the criterion that was relevant for the exclusion of each individual sequence. These tables can be downloaded in text (.txt) format.
The Pre-selection and Selection steps were developed in order to ensure that only relevant, high-quality BcR IG /TR sequence data will be included in the downstream Analytical Pipeline of TRIP.
5.3 TRIP Analytical Pipeline
Once the NGS data has been curated and filtered, it is subjected to the TRIP Analytical Pipeline (located at the “Pipeline” tab). The workflow of the analysis can be customized according to the biological context of the project.
5.3.1 Clonotype Computation
The first step of the pipeline refers to the clonotype computation. It concerns the grouping of the analyzed sequences in clonotypes, based on a set of shared immunogenetic properties.
The clustering process depends on the definition of the clonotype . TRIP provides ten different options for defining the clonotype , in order to facilitate the selection of the most relevant immunogenetic properties. If, for example, “IGHV gene and CDR3 aa sequence” is chosen as definition, all the reads expressing the same IGHV gene and identical CDR3 at the aa sequence level will be grouped together into a single clonotype .
There is also the option “Load clonotypes,” which allows to directly upload precomputed clonotypes from analyzed datasets.
The output is located at the tab “Clonotypes” and can be downloaded in text (.txt) format. The output contains a series of information regarding each individual clonotype :
-
1.
Utilized V gene and the CDR3 amino acid sequence.
-
2.
Absolute number of clustered sequences (“N”).
-
3.
Relative frequency.
-
4.
Analysis of convergent evolution referring to the number of different nucleotide sequences that encode for the CDR3 aa sequence of each given clonotype .
Each clonotype is also a link leading to a table with the immunogenetic information of all the assigned reads. At this step, each clonotype is given a unique cluster identifier (cluster ID).
Clonotype computation can provide important biological information mostly in regard to the BcR IG /TR clonality levels in a given setting. Some examples of different approaches supported by TRIP are the following:
-
1.
Frequency of the most expanded, dominant clonotype (the clonotype with the highest frequency).
-
2.
Average cumulative frequency of the “top 10” clonotypes (the ten clonotypes with the highest frequency).
-
3.
Average frequency of the abundant clonotypes (those with a frequency above a specific frequency threshold; this threshold may vary according to the aims of the project).
An example of clonality assessment using the top 10 clonotypes is illustrated in Fig. 2a.
Clonality assessment through the analysis of the top 100 clonotypes for five samples, using the either the “Clonotype computation” (a) or the “Highly similar clonotypes computation” option (b). The first three samples (namely Samples A, B, and C) display a monoclonal profile, characterized by predominance of a single clonotype with a frequency of 95%, 87.6%, and 90.9%, respectively. Sample D is oligoclonal, with multiple clonotypes exhibiting high frequency; the dominant clonotype accounts for 34.4% of the repertoire, whereas the cumulative frequency of the top 10 clonotypes is 86.4%. Finally, Sample E is polyclonal with the top 10clonotypes accounting for a very small fraction of the repertoire (2.9%). The option of merging together the “Highly similar clonotypes” resulted in an increase in the cumulative frequency of the top 10clonotypes in all samples (range 0.4–2.8%) indicating the presence of minor clonotypes exhibiting strong immunogenetic relations with the top 10 clonotypes
5.3.2 Computation of Highly Similar Clonotypes
Following the previous approach on clonotype definition, namely “V gene and CDR3 aa sequence,” two or more clonotypes would be considered as highly similar, if displaying the same CDR3 amino acid length and a low number of amino acid mismatches. TRIP allows for the grouping of highly similar clonotypes obtained at the “Clonotypes computation” step (Subheading 5.3.1). The number of allowed CDR3 aa mismatches can be either chosen manually for each individual CDR3 length or through the application of a percentage (%) threshold.
One of the most typical approaches is based on the CDR3 length and allows for a low number of aa mismatches, thus ensuring a strong connection between highly similar clonotypes:
-
1.
One aa mismatch for BcR IG /TR sequences with CDR3 lengths of up to 13 aa.
-
2.
Two aa mismatches for BcR IG /TR sequences with CDR3 lengths between 14 and 24 aa.
-
3.
Three aa mismatches for BcR IG /TR sequences with CDR3 lengths of 25 or more aa.
This process is implemented by considering the most frequent clonotype for each given CDR3 length as the reference for all the remaining clonotypes with the same CDR3 length. After merging the highly similar clonotypes, their relative frequencies are calculated accordingly.
Another parameter given by TRIP for the computation of highly similar clonotypes concerns the rearranged V gene. The application of this parameter enables the consideration of the whole variable domain of the BcR IG /TR into the clonotype grouping process, yet depends on the context of the given project.
The output files from this process are given as text (.txt) files and contain the following information:
-
1.
Cluster identifiers of the merged clonotypes (consisting of the V gene and amino acid CDR3 sequence).
-
2.
Number of sequences belonging to each merged clonotype .
-
3.
Relative frequency of each merged clonotype .
-
4.
Cluster identifiers of the clonotypes computed at the previous step (“Clonotype computation”), which formed each merged clonotype .
-
5.
Detailed information about the clonotype merging process for each individual CDR3 length.
The output files from this step can be found under the tab entitled “Highly Similar Clonotypes.”
The effect of the grouping of highly similar clonotypes on clonality assessment is given in Fig. 2b.
5.3.3 Repertoire Extraction
The next step of the analysis enables the extraction of the V, D, and J repertoires either at the gene or at the gene allele level. The V, D, or J gene repertoires are extracted from the output file of the previous step (Subheading 5.3.2) that includes all the clonotypes of the dataset (Fig. 3). Here, it is important to keep in mind that the relative frequency of each V, D, or J gene is calculated at the clonotype level rather than at the sequence level. The output of this process is provided as a text (.txt) file and contains information on the gene names, and the absolute number and relative frequency of clonotypes utilizing each specific V, D, and J gene.
At the end of this section, TRIP allows the user to choose whether the repertoire extraction will be based on the computation before or after the grouping of highly similar clonotypes. The output of this part of the pipeline can be found under the tab “Repertoires.”
5.3.4 CDR3 Length Distribution
The distribution of the CDR3 length is calculated based on the number of clonotypes corresponding to each individual length. In case the user would like to perform the analysis after the grouping of highly similar clonotypes, the results will be modified accordingly. The output is provided in the form of a table and a graph and can be found under the tab “Visualization.” Characteristic examples of CDR3 length distribution are given in Fig. 4.
The distribution of the CDR3 length in samples A–E. The x axis refers to the CDR3 length, and the y axis to the respective number of clonotypes. Samples A–C exhibited strong restrictions, in line with their monoclonal profiles. In contrast, restrictions were less prevalent in Sample D, in line with its oligoclonal clonotype repertoire. Finally, in the case of sample E, an almost Gaussian distribution of the CDR3 length is evident
5.3.5 pI Distribution
Next, the isoelectric point (pI, pH(I), and IEP) values of the CDR3 of each clonotype is extracted from the corresponding IMGT/HighV-QUEST output file, which is the pH at which the respective CDR3 carries no electrical charge or is electrically neutral. The pI of a given CDR3 is largely dependent on its amino acid composition. TRIP provides the distribution of the pI in a given dataset, based on the selection of either all or the merged clonotypes from the previous steps. A graph referring to the pI distribution can be found at the “Visualization” tab (Fig. 5).
5.3.6 Multiple Value Comparison
Different pairs of immunogenetic variables can be selected at this part of the pipeline. TRIP uses the output file from the computation of either all clonotypes or the merged clonotypes and performs comparisons between any given set of variables. The output file contains the values for each of the two selected variables and the number and relative frequency of clonotypes for each possible combination of values.
Eleven different variables that can be selected at this step include:
-
1.
V gene.
-
2.
V gene and allele.
-
3.
J gene.
-
4.
J gene and allele.
-
5.
D gene.
-
6.
D gene and allele.
-
7.
CDR3 length.
-
8.
D region reading frame.
-
9.
Molecular mass.
-
10.
pI.
-
11.
V region identity %.
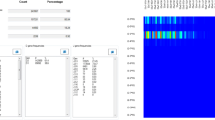
Figure 6 illustrates two examples of comparisons when using the V-gene and J-gene variables. The output files for the selected comparisons can be found at the “Multiple value comparison” tab.
Comparisons between IGHV and IGHJ gene utilization in monoclonal (Sample A) (a) versus polyclonal cases (Sample E) (b) using heatmaps. (a) A strong association between the IGHV1–8 and IGHJ4 genes is evident in Sample A, corresponding to the dominant clonotype . (b) Several associations are evident in Sample E, reflecting the polyclonal profile of this sample
5.3.7 Computation of Shared Clonotypes
In this section, TRIP scans different samples for the presence of identical clonotypes. The output file is provided in text (.txt) format with each row corresponding to a unique clonotype and each column to a different sample. Results include the absolute number of reads and the relative frequency of each clonotype in each sample (Column A: Sample id 1_Reads/Total, Column B: Sample id 1_Freq, Column C: Sample id 2_Reads/Total, Column D: Sample id 2_Freq). This type of analysis is based on the selection of either all clonotypes or just the merged clonotypes.
5.3.8 Repertoire Comparison
Similar to the comparison of clonotypes, TRIP allows the comparison of gene or gene allele repertoires (see Subheading 5.3.3), between two or more samples/datasets. The output consists of a table where each row represents a unique gene and each column a sample. Results include the absolute number and relative frequency of the clonotypes expressing each gene in every individual sample (Column A: Sample id 1_N/Total, Column B: Sample id 1_Freq, Column C: Sample id 2_N/Total, Column D: Sample id 2_Freq). Again, this type of analysis can be performed on either all clonotypes or just the merged clonotypes.
5.3.9 Clustering of CDR3 Sequences with Maximum Length Difference of One Amino Acid
As in the previous section concerning the merging of highly similar clonotypes (Subheading 5.3.2), at this point, TRIP allows for the merging of clonotypes differing by one amino acid in CDR3 length that are identical over the same length. In this case, TRIP adds one amino acid at a specified position of the shorter CDR3 resulting in the formation of two identical CDR3s. The output graph can be found at the “Visualization” tab.
5.3.10 Alignment
TRIP provides the option to align all clonotypes using the IMGT germline reference of the VDJ or VJ region at both the nucleotide and amino acid levels. An alignment table and a grouped alignment table based on the corresponding region are computed, and they are both available at the “Alignment” tab. Relevant gene alleles or a different reference sequence can be provided by the user.
5.3.11 Insert Identity Groups
At this point, TRIP enables the customization of the SHM analysis that can be applied at the next step (see Subheading 5.3.12). In detail, the user can specify the number of clonotype groups and the respective germline identity % thresholds that will be used for the SHM analysis. In certain clinical contexts, especially chronic lymphocytic leukemia (CLL), mutational categories defined by specific identity % thresholds have distinct clinical course, including responses to different treatments [21]. In that case, TRIP allows defining three distinct groups through the application of the 85–98% (see Subheading 5.2 on Selection for the application of the 85% cutoff), 98%–100%, and 100% cutoffs. The first group corresponds to “IG-mutated CLL ” (M-CLL), the second to “IG-unmutated CLL” (U-CLL), and the third to “truly IG-unmutated” CLL cases. In terms of clonotype selection, TRIP gives the user the option to perform this part of the analysis on either all or just the merged clonotypes. Figure 7 depicts the application of these identity % thresholds in a series of cases.
Relative frequency of the three IG mutational subgroups, in Samples A–E. The mutated subgroup (germline identity, GI 85–98%) was dominant in Samples A, C, and D. Truly unmutated clonotypes (GI 100%) accounted for the largest fraction of the repertoire in Sample B, indicating a different biological context. Finally, polyclonal Sample E was characterized by similar frequency levels for all mutational subgroups, perhaps due to the lack of strong selection mechanisms
This part of the analysis (“Insert identity groups”) along with the next one (“Somatic hypermutation”) apply to BcR IG datasets only, since the SHM mechanism does not operate in cells other than B cells.
5.3.12 Somatic Hypermutation Analysis
For SHM analysis, TRIP uses as reference the alignment tables produced at the Alignment step. This type of analysis can be applied to the entire dataset or only to clonotypes exhibiting either high frequency or specific immunogenetic properties.
As an output, TRIP offers information on:
-
1.
The type of nucleotide mutations and relevant amino acid changes.
-
2.
The exact position of each change, at both the nucleotide and amino acid levels.
-
3.
The topology of each change, i.e., the region of the BcR IG V domain (FR-CDR).
-
4.
The total number of clonotypes carrying each mutation.
-
5.
The frequency of each change at the gene level.
5.3.13 Logo Creation
The final step of the analysis concerns the creation of a table containing information about the frequency of each aa at each specific position of the sequence, for sequences of the same length. The region of focus is set by the user and can be either the CDR3 or the entire VDJ or VJ region. The user can also choose if TRIP will provide a frequency table for all clonotypes or just for the top N clonotypes (based on their relative frequency). Subsequently, the data in the frequency table is used for the creation of a sequence Logo. The color code used in the Logo is based on the IMGT guidelines. The output of this step can be found at the “Logo” tab.
5.3.14 Visualization
The tab “Visualization” on the TRIP interface includes all different graph types that were produced during the course of the analytical pipeline. The first graph is a bar plot of either all clonotypes or the merged clonotypes, with the option of a frequency threshold. The next graphs are pie charts of the selected V, D, and J gene repertoires. The option of applying a frequency threshold is also given here. The visualization of convergent evolution is next, with different options including a 3-D plot. Next on this tab is a pie chart and corresponding table concerning the selected identity groups for the SHM analysis along with the absolute number and frequency of clonotypes assigned to each group. The last graphs of this section are a candlestick chart for the depiction of the pI distribution and the line graph for the CDR3 distribution, below which the corresponding table is presented.
5.3.15 Overview
At this last tab of the TRIP interface, all main steps of the analysis are given, including the Preselection, Selection and Pipeline sections (Subheadings 5.1–5.3). The overview can be downloaded in pdf format. Furthermore, TRIP provides the user with the option to download all the output tables from every step of the analysis. Each table, though, can be downloaded separately, too, from its corresponding tab.
5.3.16 Dependencies
In the TRIP tool pipeline, different steps can be run independently. However, there are some dependencies:
-
1.
It is necessary to select the option “Clonotype computation” in order to apply the following types of analysis:
-
(a)
“Highly similar Clonotypes computation.”
-
(b)
“Repertoires Extraction”. In the case that the “Highly Similar Clonotypes Computation” has been selected, the repertoires will be extracted for both the total clonotypes and the merged clonotypes.
-
(c)
“Alignment” using the option “Select top N clonotypes.”
-
(d)
“Mutations” using the options “Select top N clonotypes” or “Select clonotypes separately.”
-
(e)
“Logo” using the “Select top N clonotypes” option.
-
(a)
-
2.
The “Somatic hypermutation status” is applied using the groups that have been selected using the “Insert identity groups” option.
-
3.
If both “Alignment” and “Clonotypes computation” have been selected, the cluster ID in the alignment table is the same as the one in the Clonotype table. Otherwise, all elements in the “cluster_ID” column of the alignment table will be set to 0.
-
4.
To apply “Mutations,” “Alignment” should have run previously, using the “AA or Nt” option. The Mutation table is computed based on the grouped alignment table.
References
Rawstron AC, Fazi C, Agathangelidis A et al (2016) A complementary role of multiparameter flow cytometry and high-throughput sequencing for minimal residual disease detection in chronic lymphocytic leukemia: an European research initiative on CLL study. Leukemia 30(4):929–936. https://doi.org/10.1038/leu.2015.313
Rodriguez-Vicente AE, Bikos V, Hernandez-Sanchez M et al (2017) Next-generation sequencing in chronic lymphocytic leukemia: recent findings and new horizons. Oncotarget 8(41):71234–71248. https://doi.org/10.18632/oncotarget.19525
Bolotin DA, Shugay M, Mamedov IZ et al (2013) MiTCR: software for T-cell receptor sequencing data analysis. Nat Methods 10(9):813–814. https://doi.org/10.1038/nmeth.2555
Kuchenbecker L, Nienen M, Hecht J et al (2015) IMSEQ--a fast and error aware approach to immunogenetic sequence analysis. Bioinformatics 31(18):2963–2971. https://doi.org/10.1093/bioinformatics/btv309
Thomas N, Heather J, Ndifon W et al (2013) Decombinator: a tool for fast, efficient gene assignment in T-cell receptor sequences using a finite state machine. Bioinformatics 29(5):542–550. https://doi.org/10.1093/bioinformatics/btt004
Yang X, Liu D, Lv N et al (2015) TCRklass: a new K-string-based algorithm for human and mouse TCR repertoire characterization. J Immunol 194(1):446–454. https://doi.org/10.4049/jimmunol.1400711
Aouinti S, Giudicelli V, Duroux P et al (2016) IMGT/StatClonotype for pairwise evaluation and visualization of NGS IG and TR IMGT Clonotype (AA) diversity or expression from IMGT/HighV-QUEST. Front Immunol 7:339. https://doi.org/10.3389/fimmu.2016.00339
Aouinti S, Malouche D, Giudicelli V et al (2015) IMGT/HighV-QUEST statistical significance of IMGT Clonotype (AA) diversity per gene for standardized comparisons of next generation sequencing Immunoprofiles of immunoglobulins and T cell receptors. PLoS One 10(11):e0142353. https://doi.org/10.1371/journal.pone.0142353
Bolotin DA, Poslavsky S, Mitrophanov I et al (2015) MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 12(5):380–381. https://doi.org/10.1038/nmeth.3364
Duez M, Giraud M, Herbert R et al (2016) Vidjil: a web platform for analysis of high-throughput repertoire sequencing. PLoS One 11(11):e0166126. https://doi.org/10.1371/journal.pone.0166126
Bystry V, Reigl T, Krejci A et al (2017) ARResT/interrogate: an interactive immunoprofiler for IG/TR NGS data. Bioinformatics 33(3):435–437. https://doi.org/10.1093/bioinformatics/btw634
Kotouza MT, Gemenetzi K, Galigalidou C et al (2020) TRIP - T cell receptor/immunoglobulin profiler. BMC Bioinformatics 21(1):422. https://doi.org/10.1186/s12859-020-03669-1
Gemenetzi K, Agathangelidis A, Sutton L-A et al (2018) Remarkable functional constraints on the antigen receptors of CLL stereotyped subset #2: high-throughput Immunogenetic evidence. Blood 132(Supplement 1):1839. https://doi.org/10.1182/blood-2018-99-119125
Vardi A, Vlachonikola E, Mourati S et al (2019) High-throughput B-cell immunoprofiling at diagnosis and relapse offers further evidence of functional selection throughout the natural history of chronic lymphocytic leukemia. HemaSphere 3:512. https://doi.org/10.1097/01.hs9.0000562808.48237.52
Vardi A, Vlachonikola E, Papazoglou D et al (2020) T-cell dynamics in chronic lymphocytic leukemia under different treatment modalities. Clin Cancer Res 26(18):4958–4969. https://doi.org/10.1158/1078-0432.CCR-19-3827
Vlachonikola E, Vardi A, Kastritis E et al (2018) Longitudinal T cell Immunoprofiling of patients with relapsed and/or refractory myeloma who receive Daratumumab monotherapy: a subanalysis of a phase 2 study (the REBUILD study). Blood 134(Supplement 13167):3167. https://doi.org/10.1182/blood-2019-124655
Galigalidou C, Papadopoulou A, Stalika E et al (2018) High-throughput T cell receptor (TR) repertoire analysis of virus-specific T cells: implications for T cell immunotherapy and viral infection risk stratification. Blood 132(Supplement 1):2057. https://doi.org/10.1182/blood-2018-99-118851
Gemenetzi K, Stalika E, Agathangelidis A et al (2018) Evidence for epitope-specific T cell responses in HIV-associated non neoplastic lymphadenopathy: High-Throughput Immunogenetic Evidence. Blood 132(Supplement 1):1117. https://doi.org/10.1182/blood-2018-99-118975
Lefranc MP, Giudicelli V, Duroux P et al (2015) IMGT(R), the international ImMunoGeneTics information system(R) 25 years on. Nucleic Acids Res 43(Database issue):D413–D422. https://doi.org/10.1093/nar/gku1056
Li S, Lefranc MP, Miles JJ et al (2013) IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat Commun 4:2333. https://doi.org/10.1038/ncomms3333
Chiorazzi N, Stevenson FK (2020) Celebrating 20 years of IGHV mutation analysis in CLL. HemaSphere 4(1):e334. https://doi.org/10.1097/HS9.0000000000000334
Acknowledgments
This work was supported in part by the Framework of the Hellenic Republic: Siemens Settlement Agreement, through the Hellenic Precision Medicine Network on Oncology project; the ERA-NET on Translational Cancer Research (TRANSCAN-2) acronym NOVEL project code (MIS) 5041673; and the Hellenic Foundation for Research and Innovation (HFRI) and the General Secretariat for Research and Technology (GSRT), under grant agreement No 336 (Project CLLon); the project ODYSSEAS (Intelligent and Automated Systems for enabling the Design, Simulation, and Development of Integrated Processes and Products) implemented under the “Action for the Strategic Development on the Research and Technological Sector,” funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014–2020) and @co-financed by Greece and the European Union, with grant agreement no: MIS 5002462; and the EuroClonality-NGS working group.
Disclosures: Kostas Stamatopoulos has received honoraria and research support from Abbvie, Janssen, Astra-Zeneca, and Gilead.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this protocol
Cite this protocol
Galigalidou, C., Zaragoza-Infante, L., Chatzidimitriou, A., Stamatopoulos, K., Psomopoulos, F., Agathangelidis, A. (2022). Purpose-Built Immunoinformatics for BcR IG/TR Repertoire Data Analysis. In: Langerak, A.W. (eds) Immunogenetics. Methods in Molecular Biology, vol 2453. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2115-8_27
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2115-8_27
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2114-1
Online ISBN: 978-1-0716-2115-8
eBook Packages: Springer Protocols