Abstract
Adaptive immune cells (i.e., lymphocytes of the B and T lineage) are equipped with unique antigen receptors, which collectively form a highly diverse repertoire. Within the lymphocytes, the antigen receptor diversity is created at the DNA level through recombination processes in the immunoglobulin (IG) and T cell receptor (TR) genes that encode these receptors. This gives rise to an enormous immune repertoire (a.k.a. the “immunome”) that can be studied in health and disease, both in a scientific and clinical context. In fact, the inherent distinctiveness of the IG/TR rearrangements on a per cell basis allows their usage as unique DNA fingerprints, which enables precision medicine, or for that matter “precision immunology.” The field of (fundamental and translational) research on IG/TR repertoire diversity is the topic of the Immunogenetics volume in the Methods in Molecular Biology series.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Our current understanding of the diversity of antigen receptors started with the publication on “Somatic generation of antibody diversity” by Susumu Tonegawa in 1983 [1], which resulted in the Nobel Prize in Physiology for the author in 1989. In this seminal publication, Tonegawa introduced the concept of genetic recombination mechanisms of V (variable), D (diversity), and J (joining) genes in the loci encoding the immunoglobulin (IG) chains, which—as was subsequently discovered—also applies to the T cell receptor (TR) loci. These recombinations lead to an enormous repertoire diversity of B and T lymphocytes, referred to as the “immunome.” The research into the genetics of the immune cell repertoire has been termed “immunogenetics.” Besides IG/TR gene diversity, the field of immunogenetics formally also includes diversity in the human leukocyte antigens (HLA), but this is largely beyond the scope of the current Immunogenetics volume in the Methods in Molecular Biology series.
2 Immunogenetics in the Hematology-Immunology Domain
B and T lymphocyte populations and their respective IG/TR repertoires are mostly studied in the context of immune diseases (autoimmune diseases, allergies, immune deficiencies) and immune responses (infections, inflammation, vaccinology, cancer), but also frequently in the context of hematological malignancies of immune cells (leukemias and lymphomas).
Irrespective of the application, it is important, when evaluating IG and TR repertoire diversity in B and T cell populations, to consider the repertoire data as being part of a spectrum ranging from broadly diverse (polyclonal), to restricted (oligoclonal), to dominant (clonal +/− poly/oligoclonal background) (Fig. 1). This spectrum reflects the minimal to moderate to dominant outgrowth of B or T lymphocytes of a particular specificity, which are selected based on their antigen reactivity.
Spectrum of IG/TR immune repertoire diversity, ranging from diverse (polyclonal) to highly restricted (clonal), which can be disclosed using high-throughput sequencing technologies. (Adapted from Langerak, J Immunol 2017;198:3765 [2])
Immunogenetic analysis can provide in-depth insight into the diversity of immune cells and immune responses in the context of different research questions. Additionally, the diversity or clonality of the immune repertoire can also help to address clinical and diagnostic questions. In the hematological domain, this relates to the distinction between reactive lymphoproliferations (poly- to oligoclonality) and malignantly transformed lymphocytes (clonality) or to detection of minimal residual disease of a clone upon treatment (weak clonality in background). In other areas of medicine, immunogenetic analysis can shed light on proper or defective immune responses in infected and vaccinated individuals and/or can help to distinguish between disease entities (e.g., in due time for particular autoimmune IG/TR profiles).
3 Immunogenetics Methods
Historically, immunogenetic analysis has been performed using low-resolution methodologies, such as Southern blot analysis, fragment analysis or spectratyping, and Sanger sequencing of cloned, rearranged IG/TR genes [3]. Even though these approaches enabled us to grasp the diversity of antigen receptors to some extent, they suffered from limitations in completely disclosing the depth and broadness of the IG/TR immune repertoire. The introduction of high-throughput technologies some 15 years ago allowed for a more high-resolution immune repertoire analysis via massively parallel sequencing (Fig. 2). These next-generation sequencing methods have the advantage that thousands to millions of IG/TR rearrangement sequences can be analyzed in parallel, thus approximating the true IG/TR repertoire diversity much more closely. A further development has been the introduction of single-cell sequencing technologies (Fig. 2), allowing paired analysis of different IG or TR chains at the single-cell level and the combination of immune repertoire analysis with RNA sequencing-based cell characteristics (e.g., naïve vs. memory, activated or exhausted cells).
Graphical representation of different sequencing approaches for IG/TR repertoire analysis. By means of traditional (Sanger) bulk sequencing, only the dominant immune repertoire (in green) can be identified over the background (grey), which strongly contrasts with the high-resolution output of many individual IG/TR rearrangements (represented by the different colors) through massively parallel sequencing. The additional advantage of single-cell sequencing technologies is that the high-resolution IG/TR repertoire analysis can be traced back to individual cells, which allows evaluation of paired IG or TR chains at the single-cell level and/or combination of immune repertoire and differentiation or maturation stage features
4 (Pre- and Post-)Analytical Aspects of Immunogenetics
As with any experimental method, immune repertoire analysis also entails pre-analytical, analytical, and post-analytical phases. For immune repertoire studies, the pre-analytical considerations specifically focus around the choice of sample type, nucleic acid type, IG/TR targets, etc., whereas the analytical phase relates to the pros and cons of the applied method (next-generation sequencing, quantitative PCR, droplet digital PCR). Finally, the post-analytical phase involves the readouts and tools for data analysis, but also the immuno-informatics to accurately annotate the IG/TR sequences and the bioinformatic pipelines and platforms that allow sophisticated analysis of the IG/TR data and all of their characteristic features (gene usage, CDR3, somatic mutations, clustering, and clonal evolution and competition).
In this volume of the Methods in Molecular Biology series, all of the above aspects of the pre-analytical, analytical, and post-analytical phases of IG/TR repertoire analysis are addressed in different methodological chapters that together cover a spectrum of technologies, ranging from quantitative and droplet digital PCR approaches to various NGS methodologies such as amplicon-based, capture-based, and single-cell NGS. Additionally, bioinformatic approaches are discussed that allow for extraction of IG/TR repertoire sequences from -omics data sets, i.e., RNA sequencing, whole genome sequencing, and whole exome sequencing. Finally, several novel approaches in the immunogenetic domain are covered, concerning cell-free IG/TR analysis, analysis of germline areas of the TR loci, analysis of aberrantly rearranged IG genes leading to IG translocations, and engineering of TR sequences in view of adoptive therapy.
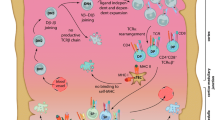
5 Immunogenetics at the Basis of Precision Immunology
Collectively, the chapters in this volume are a perfect illustration of the central position that immunogenetics has obtained in the hematology-immunology domain in both health and disease [2]. Immunogenetic profiles constitute physiological and pathophysiological signatures of cell populations, thereby allowing a more personalized approach in terms of immune responsiveness, diagnostics and classification, and even therapeutic choices [4]. This form of precision medicine involving immunogenetics could therefore best be referred to as “precision immunology” (Fig. 3). The future of immunogenetics is bright!
References
Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302(5909):575–581
Langerak AW, Brüggemann M, Davi F, Darzentas N, van Dongen JJM, Gonzalez D et al (2017) High-throughput immunogenetics for clinical and research applications in immunohematology: potential and challenges. J Immunol 198:3765–3774
Van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL et al (2003) Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98–3936. Leukemia 17:2257–2317
Arnaout RA, Prak ETL, Schwab N, Rubelt F, Adaptive Immune Receptor Repertoire Community (2021) The future of blood testing is the immunome. Front Immunol 12:626793
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this protocol
Cite this protocol
Langerak, A.W. (2022). The Advent of Precision Immunology: Immunogenetics at the Center of Immune Cell Analysis in Health and Disease. In: Langerak, A.W. (eds) Immunogenetics. Methods in Molecular Biology, vol 2453. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2115-8_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2115-8_1
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2114-1
Online ISBN: 978-1-0716-2115-8
eBook Packages: Springer Protocols