Abstract
Alternative pre-mRNA splicing can be cell-type specific and results in the generation of different protein isoforms from a single gene. Deregulation of canonical pre-mRNA splicing by disease-associated variants can result in genetic disorders. Antisense oligonucleotides (AONs) offer an attractive solution to modulate endogenous gene expression through alteration of pre-mRNA splicing events. Relevant in vitro models are crucial for appropriate evaluation of splicing modifying drugs. In this chapter, we describe how to investigate the splicing modulating activity of AONs in an in vitro skeletal muscle model, applied to Pompe disease. We also provide a detailed description of methods to visualize and analyze gene expression in differentiated skeletal muscle cells for the analysis of muscle differentiation and splicing outcome. The methodology described here is relevant to develop treatment options using AONs for other genetic muscle diseases as well, including Duchenne muscular dystrophy, myotonic dystrophy, and facioscapulohumeral muscular dystrophy.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Pre-mRNA splicing is a highly conserved process in eukaryotes that plays a role in pre-mRNA processing. Alternative splicing can diversify gene function to produce isoforms with specific functions in distinct cell types [1, 2]. Genetic variations can lead to defects in pre-mRNA splicing that cause human disease [3]. Modulation of pre-mRNA splicing can be directed to correct aberrant splicing, to skip protein coding variants, to restore the reading frame, or to prevent expression of toxic gene products. This is possible by targeting antisense oligonucleotides (AONs) toward canonical splice sites or to cis-acting regulatory elements such as cryptic splice sites or splicing silencers/enhancers [4]. Alternative splicing in skeletal muscle is abundant and essential for muscle development and function [5]. Deregulation of pre-mRNA splicing in skeletal muscle is known to be the underlying cause of multiple human myopathies [5]. Suitable in vitro and in vivo models are crucial to investigate novel splicing modulating drugs in target cells and tissues.
In vitro human skeletal muscle models can be obtained directly from muscle biopsies, or these can be generated by (trans-)differentiation of primary fibroblasts, pluripotent stem cells, or non-muscle cells with myogenic capacity like pericytes and mesoangioblasts [6,7,8]. Several protocols have been described to generate muscle progenitor cells (MPCs) derived from human patient-derived induced pluripotent stem cells (hiPSCs) using directed differentiation methods for disease modeling [9,10,11].
Here we describe how purified, expandable hiPSC-derived MPCs, generated using a transgene-free procedure [11] can be differentiated into multinucleated myotubes to test the modulating activity of AONs. These methods can be used to analyze splicing correction in vitro to develop RNA-based therapies for muscle disorders. We have used this strategy to test AONs for Pompe disease [12] and describe the methodology here in detail.
2 Materials
All cell culture work needs to be performed under sterile conditions in safety cabinets. All cell lines should be tested for mycoplasma following the manufacturer instructions (Lonza; LT07-318). Cell lines are cultured at 5% CO2 and 37 °C in humidified incubators.
2.1 Skeletal Muscle Progenitor Cell Culture
-
1.
Human MPC lines (see Note 1).
-
2.
DMEM 4.5 g/L Glucose.
-
3.
Fetal Bovine Serum.
-
4.
Penicillin/Streptomycin/Glutamine 100× (p/s/g).
-
5.
Fibroblast Growth Factor 2 (FGF2) (see Note 2).
-
6.
Sterile cell culture grade Bovine Serum Albumin (7.5% BSA).
-
7.
TrypLE™ Express Enzyme (1×), phenol red.
-
8.
Phosphate Buffered Saline (DPBS).
-
9.
Extracellular Matrix gel from Engelbreth (ECM; 1×).
-
10.
DMEM :F12.
-
11.
Insulin/Transferrin/Selenium 100×.
-
12.
DMSO.
-
13.
Freezing containers.
2.2 Cell Culture Media
-
1.
Proliferation medium: DMEM 4.5 g/L Glucose, supplemented with 10% FBS, 1× Pen/Strep, and 100 ng/ml FGF2 (added directly to plate/well).
-
2.
Differentiation medium: DMEM:F12, supplemented with 1× ITS-X and 1× Pen/Strep.
2.3 Antisense Oligonucleotide Design and Delivery
-
1.
Phosphorodiamidate morpholino oligomer (PMO) AONs (Gene Tools, LLC.)
-
2.
Endoporter (Gene Tools, LLC.)
-
3.
MilliQ filtered sterile water.
2.4 Immunofluorescence
-
1.
4% Paraformaldehyde (diluted from a 32% solution in PBS).
-
2.
0.1% Tween (diluted in PBS from a 100% solution).
-
3.
0.3% Triton-X100 (diluted in PBS from a 100% solution).
-
4.
Bovine Serum Albumin (BSA).
-
5.
Primary antibodies: Mouse-α-MYH1E (1:50, MF20 supernatant, DSHB), Rabbit-α-MYOGENIN (1:100, sc-576, Santa Cruz), Rabbit-α-MYOD (1:100, sc-304, Santa Cruz), Mouse-α-PAX7 (1:100, concentrate, DSHB).
-
6.
Secondary antibodies: Horse-α-mouse biotin (1:250, Vector Laboratories), Alexa Fluor-594-a-goat, Alexa Fluor-488-α-mouse, Alexa Fluor-594-α-rabbit, Alexa Fluor-488-α-rabbit (1:500, Invitrogen).
-
7.
Tertiary: Streptavidin 594 (1:500, Invitrogen, S-32356).
-
8.
Hoechst 33342 (1:15,000, Invitrogen, H3570).
-
9.
Nikon wide field microscope (10× and 20× objectives).
2.5 RNA Isolation, cDNA Synthesis, and Quantitative RT-PCR (RT-qPCR)
-
1.
RNA isolation kit.
-
2.
cDNA Synthesis kit.
-
3.
iTaq Universal SYBR Green Supermix.
-
4.
Hard-Shell 96-Well PCR Plates.
-
5.
Thermocycler.
-
6.
Real-time thermocycler.
-
7.
Spectrophotometer.
-
8.
Agarose.
-
9.
Ethidium Bromide.
-
10.
Primers (see Table 1).
3 Methods
3.1 Expansion, Cryopreservation, and Differentiation of MPCs
3.1.1 Expansion
-
1.
MPCs grow optimally when the confluency is between 30 and 90%. It is important to maintain this cell density throughout the expansion (see Note 3).
-
2.
Plate cells onto ECM coated plates (see Note 4). Coat plates using a solution of ECM (1:200) diluted in Proliferation medium without FGF2. Coating solution is left on the plates for 30 min at RT.
-
3.
For cell detachment, first wash plates in pre-warmed PBS at 37 °C and then treat the cells with a 1:1 pre-warmed solution of TrypLE™ Express Enzyme and PBS (3 ml for 10 cm plates) for 3–5 min at 37 °C.
-
4.
Collect cells using 5 volumes of Proliferation medium and centrifuge for 4 min at 200 × g.
-
5.
Resuspend cells using Proliferation medium, transfer to pre-coated plates (remove coating solution, do not wash), and add 100 ng/ml of FGF2 directly into the plate (see Note 5).
-
6.
Immediately transfer plates to a humidified incubator and perform cross movements to ensure appropriate cell spreading and mixing of FGF2.
3.1.2 Freeze-Thaw
-
1.
Thaw vials of MPCs in a pre-warmed water bath, transfer cell suspension slowly into 5 volumes of Proliferation medium (no FGF2), and centrifuge for 4 min at 200 × g.
-
2.
Plate cells in pre-coated plates using Proliferation medium plus 100 ng/ml of FGF2 freshly added to the cells.
-
3.
Freeze cells using Proliferation medium (plus 100 ng/ml FGF2) and 10% DMSO in 1 ml cryovials and store in freezing containers at −80 °C for 24 h (at least) prior to long-term storage in liquid nitrogen tanks.
3.1.3 Differentiation into Multinucleated Myotubes
-
1.
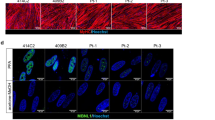
Grow cells to reach >90% confluency (avoid 100% confluency) and then switch to Differentiation medium for 4 days without refreshing (see Note 6). Wide field images of differentiated myotubes are shown in Fig. 1.
3.2 Delivery and Efficacy of Antisense Oligonucleotides in Patient-Derived Myotubes
3.2.1 Transfection
-
1.
Resuspend the PMO AONs in RNAse-free MilliQ at a concentration of 1 mM.
-
2.
Add 4.5 μl of Endoporter reagent per ml of medium directly to the cells and mix by gentle shaking (see Note 7).
-
3.
Add the desired amount of PMO AONs to the cells and mix by gentle shaking.
-
4.
Transfect AONs 1 day prior differentiation (day −1). Cells should be 60–80% confluent.
-
5.
Switch to differentiation medium (day 0).
-
6.
Leave cells to differentiate for 4 days and either collect protein or RNA or fix cells for immunofluorescence.
3.2.2 Immunofluorescence
-
1.
For immunofluorescence analysis of patient-derived myotubes, prepare cells using 48-well plates.
-
2.
Wash cells once in PBS.
-
3.
Fix cells using 4% PFA in PBS for 10 min at RT, remove and add PBS. Cells can be stored at 4 °C before proceeding.
-
4.
Wash twice in PBS for 2 min each.
-
5.
Incubate for 10 min with 0.3% Triton-X100 in PBS for permeabilization.
-
6.
Incubate for 30 min with 3% BSA, 0.1% Tween in PBS for blocking.
-
7.
Repeat washing step 4.
-
8.
Incubate with primary antibodies for 1 h at RT in 0.1% BSA, 0.1% Tween in PBS (see Note 8).
-
9.
Repeat washing step 4.
-
10.
Incubate with secondary antibodies for 45 min at RT in 0.1% BSA, 0.1% Tween in PBS.
-
11.
Repeat washing step 4.
-
12.
If biotinylated antibodies were used, incubate with tertiary for 30 min at RT in 0.1% BSA, 0.1% Tween in PBS.
-
13.
Repeat washing step 4.
-
14.
Counterstain with Hoechst nuclear staining (1:15,000) in PBS for 10 min.
-
15.
Remove and add PBS.
-
16.
Take images of five random fields with 10× or 20× lens in MYH1E-stained myotubes (see Fig. 2) to calculate the fusion index. The fusion index is determined as the percentage of nuclei present in multinucleated MYH1E-positive cells (>2 nuclei in one cell) with respect to the total number of nuclei.
3.2.3 RT-qPCR
-
1.
Harvest RNA after 4 days of differentiation using 350 μl of the lysis buffer (or the amount indicated in the first step of the RNA isolation kit) per well of a 12 well-plate.
-
2.
Purify RNA following the instructions of the preferred RNA isolation kit.
-
3.
Retrotranscribe 300–500 ng of total RNA into cDNA using a cDNA Synthesis Kit.
-
4.
Dilute cDNA samples 10× and prepare the qPCR using iTaq Universal SYBR Green Supermix.
-
5.
Amplify the cDNA of interest using a Real Time System.
-
6.
To analyze alternative splice variants in patient-derived myotubes, we normalize gene expression using each of the following four genes: MYOD, MYOG (Myogenin), LAMP1, and LAMP2 (see Note 9). Gene expression is calculated using the ΔCt method for each housekeeping gene. Thereafter, the average value of the four normalized expression values is calculated.
4 Notes
-
1.
Here we only used transgene-free derived muscle progenitor cells from hiPSCs as described in [11]. However, we anticipate that other sources of myogenic cells would also be applicable.
-
2.
The FGF2 stock powder is dissolved in 0.1% BSA (sterile cell culture-grade BSA diluted in PBS and filtered using a 0.22-μm filter) and aliquoted using tips and tubes that were coated with 0.2% BSA in PBS. The dissolved FGF2 can be stored at −80 °C. Each aliquot is used for maximally 1 week after thawing (kept at 4 °C) and 100 ng/ml is added directly to the cell culture medium every 2 days. When adding FGF2 every 2 days, this can be done without refreshing cell culture media. However, cell culture media must be refreshed every 3 days.
-
3.
MPCs spontaneously differentiate at a confluency of >90% and loose proliferative capacity in culture.
-
4.
Here we only used ECM for our studies. However, we anticipate that other coating materials can be used as well.
-
5.
We typically plate a 1/4 or 1/6 dilution of cells to get a 60–90% confluency in 2 days and 3 days, respectively, using the same plate surface area.
-
6.
There are different methods to differentiate MPCs into multinucleated myotubes [9,10,11,12]. For these studies we used DMEM:F12, 1× ITS-X, 1× p/s/g for 4 days without refreshing. Longer differentiation periods might result in cell detachment due to spontaneous contraction.
-
7.
Endoporter reagent is specifically designed for transfection of PMO AONs and was used by us in the following studies [4, 11]. Endoporter reagent does not form a complex with AONs, but it enhances endocytosis in cells. The amount of Endoporter used is independent of the concentration of PMO AONs. Other backbones might require different delivery reagents.
-
8.
The following proteins are commonly used to assess myogenic potential of differentiating MPCs across species: Myosin heavy chain (MYH1E, cytoplasmic), Myogenin (MYOG, nuclear), and MYOD (nuclear). PAX7 (nuclear) can be used to identify the muscle stem cell fraction.
-
9.
To normalize gene expression, we observed that the following genes involved in myogenesis: MYOD and MYOG; and the following involved in lysosome biogenesis: LAMP1 and LAMP2; do not change expression levels among patient and healthy donor derived myotubes. We used these genes in this study [12].
References
Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15(17):7155–7174. https://doi.org/10.1093/nar/15.17.7155
Lei Q, Li C, Zuo Z, Huang C, Cheng H, Zhou R (2016) Evolutionary insights into RNA trans-splicing in vertebrates. Genome Biol Evol 8(3):562–577. https://doi.org/10.1093/gbe/evw025
Bergsma AJ, van der Wal E, Broeders M, van der Ploeg AT, Pim Pijnappel WWM (2018) Alternative splicing in genetic diseases: improved diagnosis and novel treatment options. Int Rev Cell Mol Biol 335:85–141. https://doi.org/10.1016/bs.ircmb.2017.07.008
van der Wal E, Bergsma AJ, Pijnenburg JM, van der Ploeg AT, Pijnappel W (2017) Antisense oligonucleotides promote exon inclusion and correct the common c.-32-13T>G GAA splicing variant in Pompe disease. Mol Ther Nucleic Acids 7:90–100. https://doi.org/10.1016/j.omtn.2017.03.001
Pistoni M, Ghigna C, Gabellini D (2010) Alternative splicing and muscular dystrophy. RNA Biol 7(4):441–452. https://doi.org/10.4161/rna.7.4.12258
Baghdadi MB, Tajbakhsh S (2018) Regulation and phylogeny of skeletal muscle regeneration. Dev Biol 433(2):200–209. https://doi.org/10.1016/j.ydbio.2017.07.026
Ito N, Kii I, Shimizu N, Tanaka H, Takeda S (2017) Direct reprogramming of fibroblasts into skeletal muscle progenitor cells by transcription factors enriched in undifferentiated subpopulation of satellite cells. Sci Rep 7(1):8097. https://doi.org/10.1038/s41598-017-08232-2
Selvaraj S, Kyba M, Perlingeiro RCR (2019) Pluripotent stem cell-based therapeutics for muscular dystrophies. Trends Mol Med 25(9):803–816. https://doi.org/10.1016/j.molmed.2019.07.004
Choi IY, Lim H, Estrellas K, Mula J, Cohen TV, Zhang Y, Donnelly CJ, Richard JP, Kim YJ, Kim H, Kazuki Y, Oshimura M, Li HL, Hotta A, Rothstein J, Maragakis N, Wagner KR, Lee G (2016) Concordant but varied phenotypes among Duchenne muscular dystrophy patient-specific myoblasts derived using a human iPSC-based model. Cell Rep 15(10):2301–2312. https://doi.org/10.1016/j.celrep.2016.05.016
Hicks MR, Hiserodt J, Paras K, Fujiwara W, Eskin A, Jan M, Xi H, Young CS, Evseenko D, Nelson SF, Spencer MJ, Handel BV, Pyle AD (2018) ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs. Nat Cell Biol 20(1):46–57. https://doi.org/10.1038/s41556-017-0010-2
van der Wal E, Herrero-Hernandez P, Wan R, Broeders M, In ‘t Groen SLM, van Gestel TJM, van Ijcken WFJ, Cheung TH, van der Ploeg AT, Schaaf GJ, Pijnappel W (2018) Large-scale expansion of human iPSC-derived skeletal muscle cells for disease modeling and cell-based therapeutic strategies. Stem cell Rep 10(6):1975–1990. https://doi.org/10.1016/j.stemcr.2018.04.002
van der Wal E, Bergsma AJ, van Gestel TJM, In ‘t Groen SLM, Zaehres H, Arauzo-Bravo MJ, Scholer HR, van der Ploeg AT, Pijnappel W (2017) GAA deficiency in Pompe disease is alleviated by exon inclusion in iPSC-derived skeletal muscle cells. Mol Ther Nucleic Acids 7:101–115. https://doi.org/10.1016/j.omtn.2017.03.002
Acknowledgments
We thank Erik van der Wal for the critical reading and the revisions of this manuscript. This work was funded by Tex net, the Prinses Beatrix Spierfonds/Stichting Spieren voor Spieren (grant W.OR13-21), the Sophia Children’s Hospital Foundation (SSWO) (grant S-687 and S17-32), and Metakids (grant 2016-063).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this protocol
Cite this protocol
Herrero-Hernandez, P., Bergsma, A.J., Pijnappel, W.W.M.P. (2022). Generation of Human iPSC-Derived Myotubes to Investigate RNA-Based Therapies In Vitro. In: Arechavala-Gomeza, V., Garanto, A. (eds) Antisense RNA Design, Delivery, and Analysis. Methods in Molecular Biology, vol 2434. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2010-6_15
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2010-6_15
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2009-0
Online ISBN: 978-1-0716-2010-6
eBook Packages: Springer Protocols