Abstract
The computation of T1 maps from MR datasets represents an important step toward the precise characterization of kidney disease models in small animals. Here the main strategies to analyze renal T1 mapping datasets derived from small rodents are presented. Suggestions are provided with respect to essential software requirements, and advice is provided as to how dataset completeness and quality may be evaluated. The various fitting models applicable to T1 mapping are presented and discussed. Finally, some methods are proposed for validating the obtained results.
This chapter is based upon work from the COST Action PARENCHIMA, a community-driven network funded by the European Cooperation in Science and Technology (COST) program of the European Union, which aims to improve the reproducibility and standardization of renal MRI biomarkers. This analysis protocol chapter is complemented by two separate chapters describing the basic concept and experimental procedure.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
The computation of T1 maps from MR datasets represents an important step towards the precise characterization of kidney disease models in the small animal. Although some concepts pertaining to T1 mapping are relatively general, there are some important points that need to be observed specifically for each different acquisition strategy. In this section we will describe in more detail the three main classes of data fitting procedures corresponding to saturation recovery, inversion recovery, and variable flip angle protocols.
This analysis protocol chapter is complemented by two separate chapters describing the basic concept and experimental procedure, which are part of this book.
This chapter is part of the book Pohlmann A, Niendorf T (eds) (2020) Preclinical MRI of the Kidney—Methods and Protocols. Springer, New York.
2 Materials
2.1 Software Requirements
The method described in this chapter does require the following software tools:
-
1.
For Bruker users, the availability of a Bruker post-processing station with Bruker Paravision fitting tools (“ISA Tools”) installed, or if an independent console is not available, the scanner console. It may not always be possible or practical to alter the vendor-proposed routines to adapt to a particular use case.
-
2.
A programming environment capable of applying fitting models, such as MATLAB® including the curve fitting toolbox (MathWorks, https://www.mathworks.com/products/matlab.html), Octave (https://www.gnu.org/software/octave/), ...
-
3.
An image processing software (e.g., Image J, we recommend using Fiji, which is ImageJ with a wide range of plugins already included, https://fiji.sc/, open source), as a practical tool for the image quality check or for measuring the SNR.
-
4.
For region based approaches, analysis can be done in the R language [1].
-
5.
To speed up computation time, it may be necessary to use optimized compiled code executables or to take advantage of parallel computing.
2.2 Source Data: Format Requirements and Quality Check
2.2.1 Input Requirements
To be able to calculate T1 maps, multiple images with different repetition times, inversion times or flip angles are needed. It is necessary to have access to some scan parameters, including matrix size and number of slices, scaling information (offset and slope), and the values of the repetition times, inversion times or flip angle. Additional time delays may also be required. In the case of protocols with multiple acquisitions, the receiver gain should also be accessible. Consistency in the acquisition parameters is paramount, especially the parameter that is varied, which should sample the same interval for each animal in the study. The completeness of the data needs to be verified ahead of the processing, by checking that the matrices have the expected size and dimensionality, or in the case of multiple protocols datasets, by ensuring that each expected protocol has indeed been acquired.
2.2.2 Physiologic Motion Check
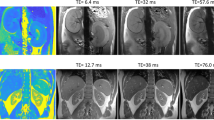
Physiological motion is an important source of image error. Because it is difficult to avoid entirely in vivo, even in the presence of triggered acquisitions, it represents an important determinant of the dataset quality. The amount of physiologic motion can be assessed visually or via quantitative estimators (see below). Frames displaying high respiratory motion may be either discarded altogether, or they may be weighted differently in the fitting procedure. In the simplest cases (especially with cartesian sampling schemes), respiratory motion can be identified as high intensity regions in the air surrounding the animal, corresponding to smeared projection of the signal along the phase-encoding direction (Fig. 1). Such motion may not be immediately visible and instead require to enhance the image windowing. Varying degrees of physiologic motion may be tolerated depending on the application. Automated methods can be put in place, to selectively remove frames where the noise level in the air region surrounding the animal exceeds a certain limit, in a retrospective fashion [2].
Illustration of physiologic respiration-based noise. Left: original image. Middle: same image with a different windowing applied. Respiratory noise is evident as a signal smeared in the phase-encoding direction (vertical) only in the image with a strong windowing applied. Different amounts of noise may be tolerated depending on the final intended use of the data. Right: image with T1 map overlay on the kidneys (color scale in ms)
2.2.3 Acquisition Geometry Check
The geometry of the acquisition also needs to be set and verified prior to processing.
-
1.
Verify the slice position and orientation to ensure that an optimal delineation of the kidneys is possible. A coronal orientation is often chosen, because it enables to have large regions of interest that depict all the anatomical regions of the kidney in a single image.
-
2.
Visually check image centering to ensure that the dataset can be exploited.
2.2.4 Acquisition Parameters Consistency Check (Multiple Scans Case Only)
Regardless of the technique which is applied, it is preferable to acquire a single protocol where the parameter of interest varies.
-
1.
Assume that the acquisition parameters are the same for all acquired images and move on to the next step.
If such a protocol is not available on the system, the same scan has to be acquired several times while varying the parameter of interest. This is the case typically for T1 mapping with variable flip angle approach.
-
2.
Check that all parameters (other than the varied parameter of interest) have stayed the same.
2.2.5 Receiver Gain Consistency Check (Multiple Scans Case Only)
-
1.
Check that the receiver gain setting is identical for each scan when merging data from separate protocols.
-
2.
If the receiver gain is not the same: contact the manufacturer to obtain receiver gain amplification tables.
-
3.
Use the receiver gain amplification tables to calculate correcting factors.
-
4.
Apply the correction factors to set each image at a common amplification level.
This exposes the user to noise compression or amplification and should only be used as a last resort.
2.2.6 Data Intensity Scaling Consistency Check (Multiple Scans Case Only)
The data should also be rescaled to the same intensity mapping. This ensures comparability of data coming from different datasets. For Bruker-generated data, it is recommended to use the absolute mapping option in the reconstruction options. If this has not been done during acquisition, additionally for Bruker-generated data, at the console, the user must:
-
1.
Duplicate the processing.
-
2.
Activate “Prototype Reconstruction”.
-
3.
Set output mapping to “User Scale”.
-
4.
Set parameters “Output Slope/Output Offset” accordingly to the setting of the first scan of the series. The setting of the first scan of the series can be checked in the “Single Parameter” parameter editor.
3 Methods
3.1 Data Exclusion
-
1.
Check that all data points have sufficient signal to noise (SNR of 5 or higher).
-
2.
If not all data points have high SNR then consider the necessity to only use data points with sufficient signal to noise (see next two steps).
-
3.
For variable repetition time protocols, eliminate the data points at low repetition times (TR) that have insufficient SNR.
-
4.
For inversion recovery protocols eliminate the data points close to the zero-crossing that have sufficient signal to noise.
3.2 Data Coregistration
To improve dataset quality, coregistration of images can be implemented to correct for respiratory motion, if such tools are available. The elastix package [3], MATLAB’s built-in imregtform function, SPM [4], or AIR [5] (among others) can be used toward that objective.
3.3 Model Fitting
With contemporary computing hardware, fast fitting of the model to the signal intensity magnitude images can be performed on each pixel of the initial dataset. A region-based approach (wherein the fitting procedure is applied to ROI averages), can also be applied, but by construction it will lack the ability to examine the local spatial variations of tissue T1.
3.3.1 Fitting Data for Saturation Recovery Experiments
-
1.
Select an appropriate fitting equation. For the VTR protocol, the following equation is applicable (under the assumption of a perfect pulse) [6]:
$$ S={S}_0\left(1-{\mathrm{e}}^{-\mathrm{TR}/{T}_1}\right)+\varepsilon $$(1)where S is the signal magnitude, S0 is the maximal available signal magnitude (including amplification factors), TR is the repetition time, T1 is the longitudinal relaxation time, and ε is an additive noise component with Rician distribution.
-
2.
If S0 is known, linearize the fitting equation. A plot of ln(1 − S/S0) against TR yields a straight line of slope − 1/T1 from which T1 can be extracted with linear regression (see Note 1).
-
3.
If the longest TR is significantly longer than T1, please refer to Note 3.
-
4.
If S0 is not known, Eq. 1 can be fitted with nonlinear regression.
-
5.
Initialize the fit with appropriate starting values for S0 (see Note 2), using the signal obtained at the longest TR (see Notes 3 and 4).
-
6.
Initialize the fit with appropriate starting values for T1 (see Note 2) using the value obtained by linear regression if applicable (see item 2).
-
7.
Perform the fit with the Levenberg-Marquardt, the trust region or the maximum likelihood algorithms (see Note 5).
3.3.2 Fitting Data for Inversion Recovery Experiments
-
1.
Select an appropriate fitting equation. For the inversion-recovery protocol, the following equation is applicable (assuming perfect pulses and absence of relaxation during pulses) [6, 7]:
$$ S=\left|{S}_0\left(1-2{\mathrm{e}}^{-\mathrm{TI}/{T}_1}\right)+\varepsilon \right| $$(2)where S is the signal magnitude, S0 is the maximal available signal magnitude (including amplification factors), TI is the inversion time, T1 is the longitudinal relaxation time, and ε is an additive noise component with Rician distribution.
-
2.
If S0 is known, linearize the fitting equation. A plot of ln((S0 − S)/(2∙S0)) against TI yields a straight line of slope −1/T1 from which T1 can be extracted with linear regression (see Note 1).
-
3.
Optionally, apply a corrective formula to account for the perturbation in recovery caused by the small angle imaging pulses (see Note 6):
$$ \frac{1}{T_1}=\frac{1}{T_1^{\ast }}+\frac{\ln \left(\cos \left(\theta \right)\right)}{\tau } $$(3) -
4.
Optionally, the following equation is also applicable (assuming perfect pulses and absence of relaxation during pulses) [2, 8,9,10,11] and adds a saturation-correcting factor (see Notes 7 and 8):
$$ S={S}_0\left(1-\beta \bullet {\mathrm{e}}^{-\mathrm{TI}/{T}_1^{\ast }}\right) $$(4) -
5.
If S0 is known, linearize fitting Eq. 4. A plot of ln(1 − S/S0) against TI yields a straight line of intercept ln(β) and of slope − 1/T1 from which T1* can be extracted with linear regression. The actual T1 is subsequently recovered from the values of \( {T}_1^{\ast } \) and β as follows:
$$ {T}_1={T}_1^{\ast}\bullet \left(\beta -1\right) $$(5) -
6.
If S0 is not known, Eqs. 2 and 4 can be fitted with nonlinear regression using S0, T1 and β as adjustable parameters and if applicable the correction of Eq. 5.
-
7.
Initialize the fit with appropriate starting values (see Note 2) for S0, using the signal obtained at the longest TI (see Notes 3 and 4).
-
8.
Initialize the fit with appropriate starting values (see Note 2) for T1 using the value obtained by linear regression if applicable (see Note 4), or using the graphical procedure outlined in Note 9.
-
9.
If the longest TR is significantly longer than T1, please refer to Note 10.
-
10.
Perform the fit with the Levenberg-Marquardt, the trust region or the maximum likelihood algorithms (see Note 5).
3.3.3 Fitting Data for Variable Flip Angle Experiments
-
1.
If only two flip angles are available, use the following simplified equation involving α1 and α2 the two flip angles used, β the ratio image obtained between the two flip angles, and TR the repetition time TR [12] to compute the T1:
$$ {T}_1=\frac{\mathrm{TR}}{\ln \left(\frac{\beta \bullet \sin \left({\alpha}_2\right)\bullet \cos \left({\alpha}_1\right)-\sin \left({\alpha}_1\right)\bullet \cos \left({\alpha}_2\right)}{\beta \bullet \sin \left({\alpha}_2\right)-\sin \left({\alpha}_1\right)}\right)} $$(6) -
2.
In the more general case, the following equation is applicable (see Notes 11 and 12):
$$ S\left(\theta \right)={S}_0\bullet \sin \left(\theta \right)\bullet \left(\left(1-{E}_1\right)/\left(1-{E}_1\bullet \cos \left(\theta \right)\right)\right) $$(7a)where
$$ {E}_1=\mathit{\exp}\left(- TR/{T}_1\right) $$(7b) -
3.
Determine T1 using linear regression on the linear form of Eqs. 7a and 7b [13]:
Plot S(θ)/sin(θ) against S(θ)/tan(θ).
-
4.
Retrieve the slope of the plot from the linear regression. This is E1 (definition in Eq. 7b).
-
5.
Compute the T1 estimate directly as −1/ln(E1).
-
6.
Retrieve the intercept of the plot from the linear regression. This is S0(1 − E1). The S0 value is estimated by dividing the intercept value that was found by 1 − E1 (using for E1 the value obtained previously).
-
7.
Alternately, determine T1 using a nonlinear fitting procedure.
-
8.
Initialize the fit with starting values (see Note 2) for S0 and T1 obtained using the linearization described previously (see Note 4).
-
9.
Perform the fit with the Levenberg-Marquardt, the trust region or the maximum likelihood algorithms (see Note 5).
3.4 Visual Display
T1 maps can be shown as gray-scaled or color-coded maps. T1 maps from the region of interest can be superimposed after coregistration on a morphological image or on an image acquired with the T1 mapping protocol to better illustrate anatomical location. Window scales, usually run from T1 = 0 to T1 = 3000 ms, should be provided with the map and should be kept constant between figures.
3.5 Quantification
The fitting algorithm is usually applied on a voxel-by-voxel basis to yield a T1 map. Mean T1 is then calculated by averaging T1 values within a region of interest. It is recommended to define a region of interest for each anatomical subregion of the kidney, as the pathology of interest can affect differently these subregions [14]. Typical ROIs are cortex, outer medulla and inner medulla, using morphological images as references to draw these ROIs. Provided sufficient spatial resolution is available, finer structures such as the outer and inner stripes of the outer medulla [15], or the corticomedullary junction [7], may be distinguished. It is also common to report T1 changes relative to baseline or to the contralateral kidney.
3.6 Results Validation
3.6.1 Evaluation of Analysis Errors and Variability Using Synthetic Data
Synthetic datasets of various complexities should be generated to test the validity of the processing solution. Artificial noise can be added to assess the stability of the fitting procedure. T1 of the numeric phantom should match the expected T1. This approach is, however, limited by the signal equation, which may or may not recapitulate the real situation complexity. Hence, the use of physical phantoms can also be recommended (see procedure outlines in the chapter by Irrera P et al. “Dynamic Contrast Enhanced (DCE) MRI-Derived Renal Perfusion and Filtration: Experimental Protocol”).
3.6.2 Comparison with Reference Values from the Literature
Relatively few studies have reported T1 values for kidney acquired in animal models at high magnetic fields (4.7 T and higher). Typical ranges for normal kidney tissue are T1 = 1000–1700 ms at magnetic field strengths of 7 or 9.4 T [14,15,16], with significant differences reported between mouse strains [16]. Acute kidney injury and unilateral nephrectomy have been reported to increase T1 values in kidney, up to T1 = 1944 ms at 7 T for severe acute injury [15] and well above T1 = 2000 ms at 9.4 T for unilateral nephrectomy [14].
4 Notes
-
1.
The accurate estimation of S0 is, not trivial. Indeed, in Eqs. 1 and 2, signal evolution is asymptotic and even at very high TR or TI values, the signal only approaches the actual S0 value. Furthermore, in these protocols, TR or TI directly affect the acquisition duration. Hence, the precision of the estimation of S0 directly affects scan duration. In the linearization approach, the available precision on S0 dictates the precision with which T1 is recovered (error propagation). Hence with this approach, increased acquisition time is needed to improve the precision of the measurement. If the duration of the experiment is a concern (dynamic studies, animal models of severe diseases, difficult anesthesia), it may be preferable to leave S0 as free parameter and to use a nonlinear fitting procedure.
-
2.
In nonlinear fitting algorithms it can be advantageous to provide starting points that are close to the final values. Indeed, this allows to decrease the number of iterations required to reach convergence, and hence can limit the computation time. Furthermore, using starting values close to the true values also helps identifying the global minimum rather than a distant local minimum. Hence, strategies can be adopted to provide as accurate as possible initial estimates for the free parameters of the nonlinear fitting procedure.
-
3.
In some cases, an initial S0 estimate for fitting initialization can be obtained from the highest signal available (at the latest saturation or inversion recovery delays for instance). This estimate will only be precise if the longest delay that is sampled is sufficiently long.
-
4.
The outlined strategies for determining reasonable starting values may not work in all cases. For instance, in the presence of high experimental noise or in the case of poorly controlled acquisition conditions, the initial estimates may prove unreliable. In this case, reasonable starting values for the T1 may be alternately obtained from the literature. Special care should be taken to select literature reference values from studies where similar magnetic field strength, protocol type and experimental disease models were used.
-
5.
Careful comparison of the performance of the fitting algorithms should be conducted on datasets obtained on the disease model and hardware setup used in the study. The various algorithms may be compared in terms of computation speed and goodness-of-fit figures of merit such as the root-mean-squared error before a final choice is made.
-
6.
In inversion recovery protocols, the T1 relaxation process is sampled several times after an inversion RF pulse using small gradient echo shots, EPI readouts or FISP readouts. Thus, the natural longitudinal recovery is perturbed by the application of the imaging RF pulses of flip angle θ and separated by a repetition interval τ. The effect can be accounted for by correcting \( {T}_1^{\ast } \), the apparent T1 resulting from the fit, using the expression given in Eq. 3 [17,18,19].
-
7.
With inversion-recovery protocols followed by multiple shots, it would be preferable to use long delays between the imaging RF pulses, to drive down the saturation effect of the fast repeating gradient echo excitation RF pulses. This is not always practical, since short delays are required to fit a large number of imaging shots within a single inversion recovery interval. This is linked to the requirement that no transverse magnetization remains at the end of an imaging shot once the subsequent imaging shot is acquired, that is, that the condition of spoiled magnetization is applicable. The generally accepted condition is thus to ensure that τ ≫ \( {T}_2^{\ast } \). Thus, although not ideal, it is acceptable to shorten the repetition time between imaging RF pulses, to reach an optimal situation where the two conflicting constraints of having both a large number of imaging shots per inversion recovery and a sufficiently good spoiling of magnetization between consecutive imaging shots are met. Similarly, it can be advantageous to work at low flip angle, to minimize the perturbation to the natural magnetization recovery relative to the ideal case when no imaging RF pulses are applied after the inversion RF pulse. This comes at a disadvantage of having a smaller signal to noise ratio. However, this may be preferable when exactness of the recovered T1 is important. In conclusion, the selection of the most appropriate fitting equation should be guided by careful examination of the specific acquisition protocol that was used, and of values of the imaging delays, flip angles, and typical expected tissue magnetic properties in the disease model at hand.
-
8.
More complete correction schemes can be applied in the case of Look–Locker protocols. In particular, the delay between the inversion RF pulse and the first imaging RF pulse may be different than the delay between subsequent sequential imaging RF pulses. The imperfections in the inversion profile may also be modelled in the equation. Expressions taking this delay into account are available. Furthermore, in inversion recovery protocols, some variants may be segmented. For instance, partial k-space segments can be acquired by groups of imaging shots at several times post inversion. The various individual shots in a segment are used to reconstruct an image at a single inversion delay (the delay between the inversion RF pulse and the half of the imaging segment). Imaging segments may be separated by arbitrary delays. Expressions are available to take into account these delays [20,21,22].
-
9.
In the case of inversion recovery protocols, a simpler method may be used to provide a reasonable estimate of T1 to be used as starting point. Indeed, the signal has the typical shape where the image magnitude first drops down to zero, then comes back up (see Fig. 2). This is due to the fact that the image magnitude is gotten from the transverse component of the magnetization, without discerning whether the magnetization is below or above the transverse plane. This offers a graphical way to determine rapidly the T1 by identifying the point at which the signal is minimal. The recovery delay at which this condition is verified is an approximation of the time when the longitudinal magnetization crosses the transverse plane. The precision of this estimate will be improved if the sampling of inversion times is dense around this point. The delay of the zero-crossing is equal to T1 × ln(2). Hence the true T1 can be identified by dividing the zero crossing delay by ln(2). This can be useful to obtain a rapid estimate of the T1 during parameter optimization. This computation can be performed very simply at each voxel by identifying the index of the signal vector where the smallest signal intensity is obtained. This strategy is, however, limited when only few points are available along the regrowth or when the signal to noise is low. Both cases can alter the accuracy of the estimation of the delay time yielding zero crossing.
-
10.
Depending on the acquisition conditions, images are available at TI (or TR) ≫ T1. In that case, Eqs. 1 and 2 collapse to S = S0. Such images can be used to normalize the data, and the nonlinear fitting procedure can be carried out on S/S0 rather than S. By suppressing the free parameter S0 from the fitting procedure, the numeric stability of the fit is improved, at the expense of the longer acquisition time required for the acquisition of the TI ≫ T1 protocol. This approach is also subject to the problem of error propagation from the S0 estimation into the T1 term exposed previously.
-
11.
The true signal behavior may differ significantly from this expression, especially if the applied flip angle is not homogeneous. In that case, a corrective, smoothly spatial-varying \( \zeta \left({B}_1^{+}\right) \) term may need to be applied to account for excitation radiofrequency field heterogeneity. The effectively obtained flip angles, θeff, are in that case a modulation of the prescribed flip angle series, θ, such that θeff = \( \zeta \left({B}_1^{+}\right) \) · θ. The evaluation of the ζ field is not trivial, and becomes especially important at higher field strengths. Post-processing methods based on large scale features in the image such as low pass filtering, or fitting of slowly spatially varying functions, can be proposed, but only extract the combined effects of transmit (\( {B}_1^{+} \)) and receive (\( {B}_1^{-} \)) heterogeneities [23]. The modulation fields can be subtracted out before performing the fit, provided that the reception field is sufficiently homogeneous. Of note, this assumption is quite difficult to validate, especially at high field strengths or when single receive RF coil elements are used as the detection circuitry.
-
12.
Alternately, a reference T1 map acquired with a Look–Locker protocol at lower spatial resolution and short acquisition time can be used to correct B1 heterogeneity. Indeed, such protocol is much less prone to B1 heterogeneity artifacts, hence the T1 it provides can be assumed to be precise [19]. This low spatial resolution reference T1 map is fed into a separate fit of Eq. 7a expressed as function of θeff = \( \zeta \left({B}_1^{+}\right) \) · θ rather than merely θ, and letting ζ as free parameter. Finally, the modulation map ζ determined in that way can be injected back into Eq. 7a, this time letting T1 and S0 as free parameters to be determined, and operating at the full spatial resolution of the VFA dataset. This approach enables a more precise quantification of T1 because the B1+ and B1− effects are effectively separated out into the ζ and the S0 maps, respectively. However, this approach is more complex and requires to acquire separate reference T1 map, increasing the overall scan time duration.
Inversion recovery plot depicting the graphical method to identify T1. An estimate of T1 is obtained by identification of the inversion time yielding the smallest available signal, and dividing this value by ln(2). Although this quantification is not optimally accurate, it may prove useful in determining sufficiently close values to serve as a good initialization point for nonlinear fitting procedures
References
Team RC (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ramaswamy R, Campbell-Washburn AE, Wells JA, Johnson SP, Pedley RB, Walker-Samuel S et al (2015) Hepatic arterial spin labelling MRI: an initial evaluation in mice. NMR Biomed 28(2):272–280
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2010) elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29(1):196–205
Huhdanpaa H, Hwang DH, Gasparian GG, Booker MT, Cen Y, Lerner A et al (2014) Image coregistration: quantitative processing framework for the assessment of brain lesions. J Digit Imaging 27(3):369–379
Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC (1998) Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr 22(1):139–152
Haacke EM, Brown RW, Thompson MR, Venkatesan R (2014) Magneticresonance imaging, physical principles and sequence design. Wiley, Hoboken, NJ
Bosch CS, Ackerman JJ, Tilton RG, Shalwitz RA (1993) In vivo NMR imaging and spectroscopic investigation of renal pathology in lean and obese rat kidneys. Magn Reson Med 29(3):335–344
Jiang K, Tang H, Mishra PK, Macura SI, Lerman LO (2018) A rapid T1 mapping method for assessment of murine kidney viability using dynamic manganese-enhanced magnetic resonance imaging. Magn Reson Med 80(1):190–199
Pastor G, Jimenez-Gonzalez M, Plaza-Garcia S, Beraza M, Reese T (2017) Fast T1 and T2 mapping methods: the zoomed U-FLARE sequence compared with EPI and snapshot-FLASH for abdominal imaging at 11.7 Tesla. MAGMA 30(3):299–307
Rajendran R, Lew SK, Yong CX, Tan J, Wang DJ, Chuang KH (2013) Quantitative mouse renal perfusion using arterial spin labeling. NMR Biomed 26(10):1225–1232
Wang JJ, Hendrich KS, Jackson EK, Ildstad ST, Williams DS, Ho C (1998) Perfusion quantitation in transplanted rat kidney by MRI with arterial spin labeling. Kidney Int 53(6):1783–1791
Ko SF, Yip HK, Zhen YY, Lee CC, Lee CC, Huang SJ et al (2017) Severe bilateral ischemic-reperfusion renal injury: hyperacute and acute changes in apparent diffusion coefficient, T1, and T2 mapping with immunohistochemical correlations. Sci Rep 7(1):1725
Cheng HL, Wright GA (2006) Rapid high-resolution T(1) mapping by variable flip angles: accurate and precise measurements in the presence of radiofrequency field inhomogeneity. Magn Reson Med 55(3):566–574
Kierulf-Lassen C, Nielsen PM, Qi H, Damgaard M, Laustsen C, Pedersen M et al (2017) Unilateral nephrectomy diminishes ischemic acute kidney injury through enhanced perfusion and reduced pro-inflammatory and pro-fibrotic responses. PLoS One 12(12):e0190009
Hueper K, Peperhove M, Rong S, Gerstenberg J, Mengel M, Meier M et al (2014) T1-mapping for assessment of ischemia-induced acute kidney injury and prediction of chronic kidney disease in mice. Eur Radiol 24(9):2252–2260
Tewes S, Gueler F, Chen R, Gutberlet M, Jang MS, Meier M et al (2017) Functional MRI for characterization of renal perfusion impairment and edema formation due to acute kidney injury in different mouse strains. PLoS One 12(3):e0173248
De Graaf R (2007) In vivo NMR spectroscopy, principles and techniques, 2nd edn. Wiley-Interscience, Hoboken, NJ
van Schie JJ, Lavini C, van Vliet LJ, Vos FM (2015) Feasibility of a fast method for B1-inhomogeneity correction for FSPGR sequences. Magn Reson Imaging 33(3):312–318
Zhang J, Chamberlain R, Etheridge M, Idiyatullin D, Corum C, Bischof J et al (2014) Quantifying iron-oxide nanoparticles at high concentration based on longitudinal relaxation using a three-dimensional SWIFT Look-Locker sequence. Magn Reson Med 71(6):1982–1988
Nkongchu K, Santyr G (2005) An improved 3-D Look-Locker imaging method for T(1) parameter estimation. Magn Reson Imaging 23(7):801–807
Henderson E, McKinnon G, Lee TY, Rutt BK (1999) A fast 3D Look-Locker method for volumetric T1 mapping. Magn Reson Imaging 17(8):1163–1171
Nkongchu K, Santyr G (2007) Phase-encoding strategies for optimal spatial resolution and T1 accuracy in 3D Look-Locker imaging. Magn Reson Imaging 25(8):1203–1214
Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PF, Gruetter R (2010) MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage 49(2):1271–1281
Acknowledgments
This chapter is based upon work from COST Action PARENCHIMA, supported by European Cooperation in Science and Technology (COST). COST (www.cost.eu) is a funding agency for research and innovation networks. COST Actions help connect research initiatives across Europe and enable scientists to enrich their ideas by sharing them with their peers. This boosts their research, career, and innovation.
PARENCHIMA (renalmri.org) is a community-driven Action in the COST program of the European Union, which unites more than 200 experts in renal MRI from 30 countries with the aim to improve the reproducibility and standardization of renal MRI biomarkers.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this protocol
Cite this protocol
Garteiser, P. et al. (2021). Analysis Protocols for MRI Mapping of Renal T1. In: Pohlmann, A., Niendorf, T. (eds) Preclinical MRI of the Kidney. Methods in Molecular Biology, vol 2216. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0978-1_35
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0978-1_35
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0977-4
Online ISBN: 978-1-0716-0978-1
eBook Packages: Springer Protocols