Abstract
Renal length and volume are important parameters in the clinical assessment of patients with diabetes mellitus, kidney transplants, or renal artery stenosis. Kidney size is used in primary diagnostics to differentiate between acute (rather swollen kidneys) and chronic (rather small kidney) pathophysiology. Total kidney volume is also an established biomarker in studies for the treatment of autosomal dominant polycystic kidney disease (ADPKD). There are several factors influencing kidney size, and there is still a debate on the value of the measured kidney size in terms of renal function or cardiovascular risk. The renal volume is most often calculated by measuring the three axes of the kidney, on the assumption that the organ resembles an ellipsoid. By default, the longitudinal and transverse diameters of the kidney are measured. In animal models renal length and volume1 are also important parameters in the assessment of organ rejection after transplantation and in determination of kidney failure due to renal artery stenosis, recurrent urinary tract infections, or diabetes mellitus. In general total kidney volume (TKV) is a valuable parameter for predicting prognosis and monitoring disease progression in animal models of human diseases like polycystic kidney disease (PKD) or acute kidney injury (AKI) and chronic kidney disease (CKD).
This chapter is based upon work from the COST Action PARENCHIMA, a community-driven network funded by the European Cooperation in Science and Technology (COST) program of the European Union, which aims to improve the reproducibility and standardization of renal MRI biomarkers. This analysis protocol is complemented by two separate chapters describing the basic concept and experimental procedure.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
Kidney size is used in primary diagnostics to differentiate between acute (rather swollen kidneys) and chronic (rather small kidney) pathophysiologies. Renal length and volume are important parameters in the clinical assessment of patients with diabetes mellitus, kidney transplants, or renal artery stenosis. Total kidney volume (TKV) is also qualified as a biomarker in studies for treatment of autosomal dominant polycystic kidney disease (ADPKD). According to the nonbinding recommendations of the FDA this biomarker can be used by drug developers for the qualified context of use in submissions of investigational new drug applications, new drug applications, and biologics license applications (www.fda.gov/media/93105/download). There are many factors governing kidney size and volume .
In patients, renal volume is probably one of the most important predictive parameters for the loss of renal function. Therefore, a determination of kidney size is recommended for patients at risk. For example in ADPKD patients <30 years with a combined renal volume >1500 mL and an estimated glomerular filtration rate (eGFR) <90 mL/min are at high risk even with otherwise normal renal function. Such patients will need renal replacement therapy within 20 years. In ADPKD patients renal volume measurements have been studied extensively and provide a method for patient stratification, monitoring of disease progression and therapeutic efficacy [1,2,3].
Also, therapeutic decisions are frequently based on the size of the kidney, and for example routinely assessed in follow-up of patients with renal stenosis or for assessment of renal transplant candidates [4, 5]. Therefore it is important to employ a measuring method that provides accurate and precise results in vivo.
In animal models renal length and volume are also important parameters in the assessment of organ rejection after transplantation and in determination of kidney failure due to renal artery stenosis, recurrent urinary tract infections, or diabetes mellitus. In general total kidney volume (TKV) is a valuable parameter for predicting prognosis and monitoring disease progression in models of polycystic kidney disease (PKD). Still, so far, no gold standard exists for renal volumetry in vivo.
The renal volume is most often calculated by measuring the three axes of the kidney, on the assumption that the organ resembles an ellipsoid. By default, the longitudinal and transverse diameters of the kidney are measured. The kidney volume is calculated according to the following approximation formula (in humans these kidney volume data correlate well with the body length and age) (see Fig. 1):
volume = length × width × average depth × 0.5.
Conventional anatomic MRI offers easy access to high quality image data. Kidney volume is reliably reproduced, and measurements can be performed with minimal bias and low inter- and intraoperator variability [6]. In the voxel-count method accurate calculation is facilitated by acquisition of multiple consecutive images sectioning the kidney. After identification of the organ boundaries, summation of all voxel volumes lying within the organ boundaries provides the total renal volume . While such an approach is highly accurate, it is also time-consuming. Transferring TKV measurement into everyday practice requires imaging techniques and protocols that are widely available while easy to employ and fast. Furthermore methods for interpretation of results are needed that are feasible and easy to apply. For this purpose open source image analysis tools are available that facilitate fast and easy determination of TKV.
For anatomical MRI of the kidney T2 weighted MRI sequences are the modality of choice. They provide excellent contrast between different tissues and for the different compartments of the kidney itself. Standard spin-echo T2 weighted imaging sequences are time-consuming due to the long repetition times TR. However, they still offer best image quality with respect to reproducibility and inter slice variability. Additionally such sequences can be modified easily to perform multiecho imaging, resulting in a set of images with different weighting that even can be used to calculate T2 maps. In this tutorial we demonstrate the applicability of a 2D T2 weighted multi echo MRI for accurate determination of kidney volume and compare different standardized TKV measurement techniques using MRI scanners developed for clinical routine imaging or dedicated to (preclinical) small animal imaging.
This chapter is part of the book Pohlmann A, Niendorf T (eds) (2020) Preclinical MRI of the Kidney—Methods and Protocols. Springer, New York.
2 Materials
2.1 Animals
These experimental protocols are tailored for mice (C57BL/6J) with a body mass of 20–30 g. Advice for adaptation to rat (Wistar, Sprague-Dawley or Lewis) is given in Subheading 4 where necessary.
2.2 Lab Equipment
-
1.
Anesthesia: For standard experiments isoflurane inhalation (CP-Pharma, Baxter) provides robust anesthesia for up to 2 h with comparatively little side-effects on renal physiology. For a detailed description and discussion of the anesthesia please refer to the chapter by Kaucsar T et al. “Preparation and Monitoring of Small Animals in Renal MRI.”
-
2.
Gases: O2 or compressed air, as delivering system for evaporated isoflurane. Besides air for use with pulse oximetry systems for monitoring blood oxygenation, O2 gas is preferred during the experiment on diseased animals.
-
3.
Devices for physiological monitoring ECG, temperature and respiration, to trigger the image acquisition: for example SAI (Model 1030, SAII, Stony Brook, NY, US ).
2.3 MRI Hardware
The general hardware requirements for renal 1H MRI on mice and rats are described in the chapter by Ramos Delgado P et al. “Hardware Considerations for Preclinical Magnetic Resonance of the Kidney” (open-access). The technique described in this chapter was tailored for a 9.4 T MR system (Biospec 94/20, Bruker Biospin, Ettlingen, Germany) but advice for adaptation to other field strengths and systems (e.g., 4.7 T Varian and 3 T Siemens Skyra human MR scanner using a wrist RF coil (for signal reception) or knee RF coil (transmit-receive)) is given where necessary.
With preclinical MRI systems volume RF coils covering the entire mouse or rat bodies can be used for signal transmission and reception. However, if needed signal-to-noise ratio (SNR) can be elevated by using dedicated surface receive RF coils (i.e., mouse heart four-element surface RF coil or rat heart four-element surface RF coil) in combination with linear polarized transmit only volume RF coils.
No other special or additional hardware is required.
2.4 MRI Protocols
For anatomical MRI of the kidney T2-weighted MRI sequences are the modality of choice. Accelerated imaging techniques are available on all MRI systems. On Bruker systems they are identified by acronyms “RARE” or “turboRARE” (for rapid acquisition relaxation enhanced). On Philips and Siemens scanners such sequences usually are denoted “FSE” or “TSE” (for fast spin echo or turbo spin echo).
2.5 Image Analysis Tools
MRI data can be analyzed easily by manual planimetry or by calculating TKV from length and width measurements with different standardized equations2 (the “Traditional Ellipsoid,” the “Mayo Ellipsoid,” and the “Mid-slice Method”). For this we recommend employing the open source imaging tools ImageJ or IcY:
-
1.
ImageJ (https://imagej.nih.gov/ij/) and the Versatile Wand Tool (https://imagej.nih.gov/ij/plugins/versatile-wand-tool/index.html).
- 2.
For providing the ex vivo gold standard, kidney volumes can be additionally measured post mortem, using the fluid displacement method.
3 Methods
Renal volumes can be calculated in several ways, using the ellipsoid formula or the voxel-count method. For the ellipsoid formula calculation, the length is determined on the sagittal scans. The width and thickness will be measured at the hilum on the transverse scans. Width can also be measured at the largest transverse diameter. Both volume-hilum and volume-maximum will be calculated. Volume measurements using the ellipsoid formula can easily be done in less than 2 min. In most clinical studies, the ellipsoid method is commonly applied for renal volume assessment. With this method, it is assumed that the kidney resembles an ellipsoid structure. This leads to systematic underestimation of the renal volume . In fact the kidney is not a true ellipsoid structure.
With the voxel-count method, the volumes of all voxels within the boundary of the kidney are summated, thus giving the true total volume of the kidney so that obtaining inaccurate results is highly unlikely. For the voxel-count method, the kidney has to be segmented manually. Segmentations can be done by tracing the boundaries of the kidney on each slice. The total renal volume will then be calculated by summation of all voxel volumes lying within the boundaries of the kidney. Partial volume effects, which occur if voxels contain both kidney and surrounding tissue, could lead to overestimation of the kidney volume , if such voxels are included within the boundaries of the kidney. To avoid such an overestimation, the segmentation line can be drawn halfway along the change in signal intensity between the kidney and surrounding tissues. Semiautomatic segmentation techniques, such as region-growing, can save time. However, such methods are not really practical to use for most available software. Neighboring tissues with very similar signal intensity still have to be separated manually. Fat within the kidneys might perturb the segmentation of the boundaries due to fat-water chemical shift artifacts when using region-growing segmentation technique, leading to an underestimation of the total volume . Semiautomatic segmentation techniques are also challenging to perform on images obtained with accelerated T2 weighted MRI sequences. While accelerated T2 weighted imaging yields good results when organ morphology is considered, signal to noise ratio fluctuations between the individual slices due to spatial changes in the noise amplification intrinsic to parallel imaging techniques cannot be entirely prevented. For this reason, selection of threshold values and propagation has to be done individually for each slice and is a source for investigator bias and experimental error. Newer segmentation techniques, like automatic contour detection, might be an option in future software implementations.
Calculating renal volume from both coronal and sagittal scans can help eliminating differences due to aberrations in slice positioning.
Furthermore there is a simplified Mid-Slice Technique for MRI. In this technique, the renal volume is calculated from the area of a single middle slice image of the kidney multiplied by the number of slices. The kidney volumes correlate well with stereology and have high reproducibility comparable with manual planimetry. However, when calculating single kidney volumes, both the mid-slice technique and the ellipsoid formula are less accurate than stereology and manual or semiautomatic planimetry. Although significantly faster than manual tracing for calculating kidney volume , this technique is slower than the standard ellipsoid method. Volume estimates are based on a multiplier linked to the hypothesis that the shape of the kidney is ellipsoidal.
All these approaches rely on geometrical assumptions, that might not be true.
3.1 MR Protocol Setup
3.1.1 Multislice Multiecho Sequence for T2 Imaging
-
1.
Load the 2D multislice multiecho sequence (MSME). (preferred see Note 1)
-
2.
Set the shortest echo time (TE) and echo spacing (ΔTE) possible, under the condition that fat and water are in phase (see Note 2). The last TE should be close to the largest expected T2(*) in the kidney multiplied by 1.5 (see Note 3). The aim is to acquire at least five echo images. Consider increasing the acquisition bandwidth and using half Fourier acceleration to shorten the first TE and ΔTE (see Note 4).
-
3.
Choose the shortest possible repetition time (TR) for good signal-to-noise per time (SNR/t) efficiency. TR will be limited by the length of the echo train and the number of slices you acquire.
-
4.
Adapt the flip angle (FA) to the TR and T1 in order to achieve the best possible SNR. Use the Ernst angle αE = arccos (exp (−TR/T1)) as a good starting value. Then try a few smaller and larger FAs and determine the optimal FA experimentally by comparing the measured SNRs.
-
5.
Set a high acquisition bandwidth (BW) to shorten ΔTE, while keeping an eye on the SNR, which decreases with the square root of BW. Low SNR may be balanced out with averaging (see Note 5).
-
6.
Enable fat saturation. On ultrahigh field systems this works well to avoid fat signal overlaying the kidney due to chemical shift. At lower field strengths it might work less efficient.
-
7.
Enable the respiration trigger (per phase step or per slice). This is essential to reduce motion artifacts (see also Note 6), reduce motion blurring and unwanted intensities variations among the images acquired with different TEs.
-
8.
Choose as phase-encoding direction the L-R direction and adapt the geometry so that the FOV in this direction includes the entire animal (approx. 40 mm).
-
9.
Use frequency encoding in head-feet (rostral-caudal) direction to avoid severe aliasing. Adjust the FOV to your needs keeping in mind that in this direction the FOV can be smaller than the animal and a smaller FOV permits a smaller acquisition matrix, and in turn a shorter echo-spacing.
-
10.
Use an appropriate slice thickness, typically around 1.0 mm.
-
11.
Use high in-plane resolution that the SNR allows, typically between 100 and 200 μm. Zero-filling in phase encoding direction can be helpful to speed up acquisition. One may use half Fourier in read direction (asymmetric echo) to further shorten the first TE, if very short T2* (<5 ms) can occur. Reducing the excitation pulse length to below 1 ms would then also help to shorten TE.
-
12.
A spin echo sequence (MSME) with an echo time of >20 ms is very sensitive for instabilities of your system. If the system is not stable for any reason, this can often be observed directly at the time signal.
-
13.
For examples of specific parameter sets please see Notes 9–13.
3.2 In Vivo MR Imaging
After obtaining scout images in the x, y, and z planes, T2-weighted MRI should be performed in sagittal and coronal orientation.
When selecting a certain type of MRI sequence and its parameters, an optimum should be established between spatial resolution, signal-to-noise ratio, and scan duration. Accuracy of the volume measurements can be enhanced by reducing slice thickness and/or choosing an additional section orientation transverse to the major axis of the kidney, mostly in axial orientation.
While theoretically beneficial for discrimination of kidney size differences, measurements in multiple orthogonal planes across the kidney prolong examination times and increase the time required for segmentation , thus hampering both clinical and preclinical practice. This holds also true for application of sophisticated high resolution T2 weighted 3D imaging techniques instead of standard (multislice) 2D T2 weighted MRI covering the entire kidney. Especially in preclinical studies of diseased animals, it is often more appropriate to use solely a single slice orientation 2D MRI approach to gather standardized images in minimum time (i.e., to reduce length of anesthesia). If reduction of imaging time is of particular importance, accelerated imaging techniques are recommended. Such sequences should be available on all MRI systems. On Bruker systems they are identified by acronyms “RARE” or “turboRARE” (for rapid acquisition relaxation enhanced). On Siemens scanners such sequences usually are denoted “FSE” or “TSE” (for fast spin echo or turbo spin echo). In these measurement techniques acceleration is facilitated mainly by recording of multiple lines of k-space, that is, performing multiple phase-encoding steps on the echo train.
3.2.1 Scanner Adjustments and Anatomical Imaging
-
1.
Acquire a fast pilot scan to obtain images in the three orthogonal planes x, y, and z.
-
2.
Acquire anatomical images in several oblique orientations to facilitate planning a coronal slice orientation with regard to the long axis of the kidney, as described in the chapter by Pohlmann A et al. “Essential Practical Steps for MRI of the Kidney in Experimental Research.”
-
3.
Perform localized shimming on the kidney as described in the chapter by Pohlmann A et al. “Essential Practical Steps for MRI of the Kidney in Experimental Research.”
3.2.2 Morphometric MR Imaging
-
1.
Load the MSME sequence , adapt the slice orientation to provide a coronal or axial view with respect to the kidney (in scanner coordinates this is double-oblique). (caveat see Note 8)
-
2.
In the monitoring unit set the trigger delay so that the trigger starts at the beginning of the expiratory plateau (no chest or diaphragm motion) and the duration such that it covers the entire expiratory phase, that is, until just before inhalation starts (1/2 to 2/3 of breath-to-breath interval) (see Note 7).
-
3.
Adapt TR to be a little shorter (about 100 ms) than the average respiration interval that is displayed on the physiological monitoring unit.
-
4.
Run the MSME scan. Example images are shown in Figs. 2, 3, and 4.
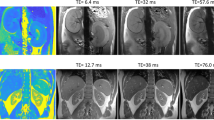
T2 weighted image of mouse kidneys in coronal view. For the quantification of kidney volume, respiratory triggered, fat-saturated T2-weighted turbo spin echo (TSE) sequences were acquired in axial and coronal planes, covering both kidneys. The coronal plane was adjusted to the long axis of both kidneys. Scan parameters were: TR/TE = 1500/33 ms, averages = 2, matrix = 256x256, field of view = 35x35 mm2, slice thickness = 1 mm
A demonstration of the volume changes that can be expected in pathophysiological scenarios is given in Fig. 4.
4 Notes
-
1.
A 3D version of the turboRARE sequence is also available, which allows thinner slices with better SNR, but tends to be too slow for most in vivo applications.
-
2.
The fat signal has a slightly different Larmor frequency (fat-water shift: Δf = 3.5 ppm × Larmor frequency or 146 Hz/T, for example: Δf = 220 Hz at 1.5 T) than the water signal. The faster precession of the fat protons means that with increasing time the fat and water signal fractions within a voxel are sometimes in phase (signals add up) and sometime out of phase (signals subtract). This can lead to an unwanted fat-water shift induced signal intensity modulation along the exponential signal decay curve. This is mostly relevant for diseased kidneys with increased fat content (e.g., diabetes), but we recommend to generally take this into account, that is, also for healthy kidneys. The TEs at which fat and water are in phase depend on the field strength: TE [ms] = n × (6.7069/B0[T]).
-
3.
When establishing the MR technique you need to define an SNR acceptance threshold for the image with the shortest TE. The aim is to have at least three (better five or more) number of echoes with an SNR > 5. This threshold will depend on the expected T2(*) values, which in turn depends on parameters like the magnetic field strength, shim quality, and tissue properties (pathology). Example: For a rat imaged at 9.4 T using a four-element rat heart array receive surface RF coil together with a volume RF resonator for excitation in combination with interventions leading to strong hypoxia, an SNR > 60 was needed.
-
4.
For rats increase the FOV to the body width and keep the matrix size the same. The relative resolution is then identical and the SNR should also be similar, because the smaller mouse RF coil provides better SNR.
-
5.
A good starting point is to use the same relative resolution as for rats. For this, reduce the FOV to the mouse body width and keep the matrix size the same.
-
6.
If no specific animal holder is used, it is preferable to position the animals in left or right decubitus position to keep the bowels away from the kidneys to mitigate susceptibility artifacts.
-
7.
You must monitor the respiration continuously throughout the entire experiment and if necessary adapt the TR accordingly.
-
8.
Acquiring more echoes with smaller echo spacing will be beneficial because it improves the fitting when calculating a T2 map (see Fig.3), but keep in mind the specific absorption rate (SAR) associated with sending many 180° RF pulses in a short time could heat up the tissue. This will usually not be detectable via a rectal temperature probe, but measurements with a temperature probe placed in the abdomen next to the kidney showed that significant temperature increases are possible with a multispin echo sequence .
-
9.
Ellipsoid based calculation: Example parameters for a 300 g rat at 9.4 T (Bruker small animal system): TR = [respiration interval] -100 ms; receiver bandwidth = 50 kHz; number of echoes = 12; first echo = 10.0 ms; echo spacing 10.0 ms; TE = 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, 120 ms; averages = 1; slice orientation = coronal to kidney; frequency encoding = head-feet; FOV = (38.2 × 48.5) mm; matrix size = 169 × 115 zero-filled to 169 × 215; resolution = (0.226 × 0.421) mm; 1 slice with 1.4 mm thickness; fat suppression = on; respiration trigger = per slice; acquisition time = 55–75 s (with triggering under urethane anesthesia).
-
10.
Ellipsoid based calculation: Example parameters for a 30 g mouse at 4.7 T (Agilent small animal system): TR = [respiration interval] − 100 ms; receiver bandwidth = 50 kHz; number of echoes = 12; first echo = 10.0 ms; echo spacing 10.0 ms; TE = 10, ..., 120 ms; averages = 1; slice orientation = coronal to kidney; frequency encoding = head-feet; FOV = (30 × 30) mm; matrix size = 128 × 128; resolution = (0.230 × 0.230) mm; 1 slice with 1.0 mm thickness; fat suppression = on; respiration trigger = on; acquisition time = 3.5–9.0 min (with triggering under isoflurane anesthesia).
-
11.
Ellipsoid based calculation: Example parameters for a 300 g rat at 3.0 T (Siemens Skyrafit, a clinical system): Animal position: Right decubitus; Coil: Knee; TR = [respiration interval] − 500 ms; receiver bandwidth = 399 Hz/pixel; number of echoes = 12; first echo = 10.0 ms; echo spacing 10.0 ms; TE = 10, ..., 120 ms; averages = 2; slice orientation = axial; frequency encoding = left-right; FOV = (120x60) mm; matrix size = 256 × 128; resolution = (0.470 × 0.470) mm; 1 slice with 2.0 mm thickness; fat suppression = on; respiration trigger = off; acquisition time ~ 2 min. If no specific animal holder is used, it is preferable to position the animals in left of right decubitus position to keep the bowels away from the kidneys to mitigate susceptibility artifacts.
-
12.
Planimetry based calculation: Example parameters for a 300 g rat at 9.4 T (Bruker small animal system): TR = 1700 ms; receiver bandwidth = 50 kHz; number of echoes = 12; first echo = 10.0 ms; echo spacing 10.0 ms; TE = 10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, 120 ms; averages = 1; slice orientation = coronal to kidney; frequency encoding = head-feet; FOV = (38.2 × 48.5) mm; matrix size = 169 × 115 zero-filled to 169 × 215; resolution = (0.226 × 0.421) mm; 13 slices at 1 mm thickness; fat suppression = on; respiration trigger = per slice; acquisition time = 220–300 s (with triggering under isoflurane anesthesia).
The following images should be acquired: coronal and transverse T2-weighted fast spin-echo sequence (TSE) and coronal T1-weighted TSE.
The following images should be acquired: coronal and transverse T2-weighted fast spin-echo sequence (TSE) and coronal T1-weighted TSE.
The parameters for coronal position TSE can be as follows: slice thickness, 2.0 mm; slice interval, 0.2 mm.
For T1 weighted turbo spin-echo (TSE): repetition time (TR), 650 ms; echo time (TE), 10 ms; field of view (FOV), 120 mm × 120 mm; band width, 250 Hz/Px; matrix, 256 × 256; and number of excitations (NEX), 4.0;
For T2 weighted TSE you can use: TR, 3460 ms; TE, 35 ms; FOV, 120 mm × 120 mm; band width, 250 Hz/Px; matrix, 256 × 256; and NEX, 4.0.
The parameters for the transverse position T2 TSE can be as follows: slice thickness, 2.0 mm; slice interval, 0.2 mm; TR, 650 ms; TE, 10 ms; FOV, 120 mm × 120 mm; band width, 250 Hz/Px; matrix, 256 × 256; and NEX, 4.0.
Shimming is particularly important, since macroscopic magnetic field inhomogeneities shorten T2*, but provide no tissue specific information—rather they overshadow the microscopic T2* effects of interest and hinder quantitative intra- and intersubject comparisons. Shimming should be performed on a voxel enclosing only the kidney using either the default iterative shimming method or based on previously recorded field maps (recommended).
-
13.
If the height adjusted total kidney volume (HtTKV) must be calculated it can be done by measuring the kidneys in three axes. To calculate HtTKV a structural MRI or at least CT scan is required. The height of the patient is also required for the calculation. The total kidney volume (TKV) is determined using an ellipsoid equation. This is clinically validated for patients with polycystic disease, with bilateral and diffuse small to medium sized cysts. The ellipsoid equation requires only three measurements from both kidneys:
-
Kidney length.
-
Kidney width.
-
Kidney depth.
-
If you are viewing images on PACs or any DICOM-Viewer it is easiest to calculate the HtTKV with all planes of the kidney visible at the same time. To do this, you mostly have to select “MPR” under the available “View Options” in the specific menu.With this you can view the kidney in the coronal, sagittal, and axial planes. The maximum length of the kidney should be measured in the sagittal plane.
The width and depth can be measured in the axial plane in a slice that shows the greatest kidney width and depth.Repeat this for both kidneys. Once you have all six measurements and the patient’s height, you can calculate HtTKV or use one of the available online calculators for kidney volume .
References
King BF, Reed JE, Bergstralh EJ, et al (2000) Quantification and longitudinal trends of kidney, renal cyst, and renal parenchyma volumes in autosomal dominant polycystic kidney disease. 11:1505–1511
Sise C, Kusaka M, Wetzel LH et al (2000) Volumetric determination of progression in autosomal dominant polycystic kidney disease by computed tomography. Kidney Int 58:2492–2501
Grantham JJ, Torres VE (2016) The importance of total kidney volume in evaluating progression of polycystic kidney disease. Nat Rev Nephrol 12:667–677
Hederström E, Forsberg L (1985) Kidney size in children assessed by ultrasonography and urography. Acta Radiol Diagn (Stockh) 26:85–91
Ferrer FA, McKenna PH, Bauer MB et al (1997) Accuracy of renal ultrasound measurements for predicting actual kidney size. J Urol 157:2278–2281
Sharma K, Caroli A, Quach LV et al (2017) Kidney volume measurement methods for clinical studies on autosomal dominant polycystic kidney disease. PLoS One 12:e0178488
Acknowledgments
This chapter is based upon work from COST Action PARENCHIMA, supported by European Cooperation in Science and Technology (COST). COST (www.cost.eu) is a funding agency for research and innovation networks. COST Actions help connect research initiatives across Europe and enable scientists to enrich their ideas by sharing them with their peers. This boosts their research, career, and innovation.
PARENCHIMA (renalmri.org) is a community-driven Action in the COST program of the European Union, which unites more than 200 experts in renal MRI from 30 countries with the aim to improve the reproducibility and standardization of renal MRI biomarkers.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2021 The Author(s)
About this protocol
Cite this protocol
Müller, A., Meier, M. (2021). Assessment of Renal Volume with MRI: Experimental Protocol. In: Pohlmann, A., Niendorf, T. (eds) Preclinical MRI of the Kidney. Methods in Molecular Biology, vol 2216. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0978-1_21
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0978-1_21
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0977-4
Online ISBN: 978-1-0716-0978-1
eBook Packages: Springer Protocols