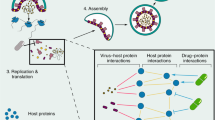
Abstract
SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) is a novel virus in the family of Coronaviridae. The virus causes COVID-19, an infectious disease. SARS-CoV-2 provides novel clinical, immunological, and pathological features, compared to other established coronaviruses, such as the Middle East respiratory syndrome (MERS-CoV) and severe acute respiratory syndrome (SARS-CoV). There is no indication of successful antiviral treatment or vaccination at this stage. Several computational methods have been used to quickly understand the pathogenesis of viruses and to classify antiviral medications. Protein–protein interaction networks allow us to understand pathogenic pathways that contribute to the disease, infection, and development and to translate this knowledge to effective diagnostics and therapeutic strategies. The conventional approach to drug intervention and treatment interventions started to evolve with the complexity of dependence and drug pathways. Identifying drug–target interactions cannot only minimize drug production cycles and costs but can also strengthen awareness of how the future drugs and targets are identified. The purpose of a pharmaceutical network is to classify multi-target compounds focused at different protein groups involved in disrupted complexes. This promotes a network-centered viewpoint on drug action via the mapping of the goal network and offers a fresh insight into the role of polypharmacology in the drug operation. In this chapter, we shall address the available experiments and computations of protein–protein interaction methods to identify SARS-CoV-2 drugs.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Boezio B, Audouze K et al (2017) Network-based approaches in pharmacology. Wiley Online Library, Weinheim
Legrain P, Wojcik J, Gauthier JM (2001) Protein-protein interaction maps: a lead towards cellular functions. Trends Genet 17:346–352
Cho S, Park SG, Lee DH, Park BC (2004) Protein-protein interaction networks: from interactions to networks. J Biochem Mol Biol 37:45–52
Ezkurdia I, Bartoli L, Fariselli P et al (2009) Progress and challenges in predicting protein-protein interaction sites. Brief Bioinform 10:233–246. https://doi.org/10.1093/bib/bbp021
Shoemaker BA, Panchenko AR (2007) Deciphering protein–protein interactions. Part II. Computational methods to predict protein and domain interaction partners. PLoS Comput Biol 3:e43. https://doi.org/10.1371/journal.pcbi.0030043
Ruffner H, Bauer A, Bouwmeester T (2007) Human protein-protein interaction networks and the value for drug discovery. Drug Discov Today 12:709–716
Jin L, Wang W, Fang G (2014) Targeting protein-protein interaction by small molecules. Annu Rev Pharmacol Toxicol 54:435–456. https://doi.org/10.1146/annurev-pharmtox-011613-140028
Kitano H (2002) Systems biology: a brief overview. Science 295:1662–1664
Bruggeman FJ, Westerhoff H (2007) The nature of systems biology. Trends Microbiol 15:45–50
Westerhoff HV, Palsson BO (2004) The evolution of molecular biology into systems biology. Nat Biotechnol 22:1249–1252
Butcher EC, Berg EL, Kunkel EJ (2004) Systems biology in drug discovery. Nat Biotechnol 22:1253–1259
Hood L, Heath JR, Phelps ME, Lin B (2004) Systems biology and new technologies enable predictive and preventative medicine. Science 306:640–643
Hucka M, Finney A, Sauro HM et al (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19:524–531. https://doi.org/10.1093/bioinformatics/btg015
Patil A, Kinoshita K, Nakamura H (2010) Hub promiscuity in protein-protein interaction networks. Int J Mol Sci 11:1930–1943. https://doi.org/10.3390/ijms11041930
Jhoti H (2001) High-throughput structural proteomics using x-rays. Trends Biotechnol 19:S67–S71
Protein structure determination in solution by NMR spectroscopy. https://www.jbc.org/content/265/36/22059.short. Accessed 27 Sep 2020
Wagner K, Racaityte K, Unger KK et al (2000) Protein mapping by two-dimensional high performance liquid chromatography. J Chromatogr A 893:293–305. https://doi.org/10.1016/S0021-9673(00)00736-6
Sydor JR, Scalf M, Sideris S et al (2003) Chip-based analysis of protein-protein interactions by fluorescence detection and on-chip immunoprecipitation combined with μLC-MS/MS analysis. Anal Chem 75:6163–6170. https://doi.org/10.1021/ac034258u
Baskaran K, Duarte JM, Biyani N et al (2014) A PDB-wide, evolution-based assessment of protein-protein interfaces. BMC Struct Biol 14:1–11. https://doi.org/10.1186/s12900-014-0022-0
Berggård T, Linse S, James P (2007) Methods for the detection and analysis of protein-protein interactions. Proteomics 7:2833–2842
Wessels HJCT, Vogel RO, van den Heuvel L et al (2009) LC-MS/MS as an alternative for SDS-PAGE in blue native analysis of protein complexes. Proteomics 9:4221–4228. https://doi.org/10.1002/pmic.200900157
Muronetz VI, Sholukh M, Korpela T (2001) Use of protein-protein interactions in affinity chromatography. J Biochem Biophys Methods 49:29–47. https://doi.org/10.1016/S0165-022X(01)00187-7
Kukar T, Eckenrode S, Gu Y et al (2002) Protein microarrays to detect protein-protein interactions using red and green fluorescent proteins. Anal Biochem 306:50–54. https://doi.org/10.1006/abio.2002.5614
Kanno A, Ozawa T, Umezawa Y (2011) Detection of protein-protein interactions in bacteria by GFP-fragment reconstitution. Methods Mol Biol 705:251–258. https://doi.org/10.1007/978-1-61737-967-3_15
Luban J, Goff SP (1995) The yeast two-hybrid system for studying protein-protein interactions. Curr Opin Biotechnol 6:59–64. https://doi.org/10.1016/0958-1669(95)80010-7
Walhout AJM, Vidal M (2001) High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24:297–306. https://doi.org/10.1006/meth.2001.1190
Coates P, Hall P (2003) The yeast two-hybrid system for identifying protein-protein interactions. J Pathol 199:4–7. https://doi.org/10.1002/path.1267
Mehla J, Caufield JH, Uetz P (2015) The yeast two-hybrid system: a tool for mapping protein-protein interactions. Cold Spring Harb Protoc 2015:425–430. https://doi.org/10.1101/pdb.top083345
Legrain P, Selig L (2000) Genome-wide protein interaction maps using two-hybrid systems. FEBS Lett 480:32–36
van Criekinge W, Beyaert R (1999) Yeast two-hybrid: state of the art. Biol Proced Online 2:1–38. https://doi.org/10.1251/bpo16
Skrabanek L, Saini HK, Bader GD, Enright AJ (2008) Computational prediction of protein-protein interactions. Mol Biotechnol 38:1–17
Tong AHY, Drees B, Nardelli G et al (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295:321–324. https://doi.org/10.1126/science.1064987
Pitre S, Alamgir M, Green JR et al (2008) Computational methods for predicting protein-protein interactions. Adv Biochem Eng Biotechnol 110:247–267
Martin EPG, Bremer EG, Guerin MC et al (2004) Analysis of protein/protein interactions through biomedical literature: text mining of abstracts vs. text mining of full text articles. In: Lecture notes in artificial intelligence (subseries of Lecture notes in computer science). Springer, Berlin, pp 96–108
Chowdhary R, Zhang J, Tan SL et al (2013) PIMiner: a web tool for extraction of protein interactions from biomedical literature. Int J Data Min Bioinform 7:450. https://doi.org/10.1504/IJDMB.2013.054232
de Bruijn B, Martin J (2002) Getting to the (c)ore of knowledge: mining biomedical literature. Int J Med Inform 67:7
Shatkay H, Feldman R (2003) Mining the biomedical literature in the genomic era: an overview. J Comput Biol 10:821–855
Chaussabel D (2004) Biomedical literature mining: challenges and solutions in the “omics” era. Am J Pharmacogenomics 4:383–393
Jensen LJ, Saric J, Bork P (2006) Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet 7:119–129
Zhou D, He Y (2008) Extracting interactions between proteins from the literature. J Biomed Inform 41:393–407
Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM (2005) Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol 6:1–12. https://doi.org/10.1186/gb-2005-6-5-r40
Krallinger M, Leitner F, Valencia A (2010) Analysis of biological processes and diseases using text mining approaches. Methods Mol Biol 593:341–382
Soong TT, Wrzeszczynski KO, Rost B (2008) Physical protein-protein interactions predicted from microarrays. Bioinformatics 24:2608–2614. https://doi.org/10.1093/bioinformatics/btn498
Chua HN, Sung WK, Wong L (2006) Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics 22:1623–1630. https://doi.org/10.1093/bioinformatics/btl145
Chua HN, Sung WK, Wong L (2007) Using indirect protein interactions for the prediction of Gene Ontology functions. BMC Bioinformatics 8:1–13. https://doi.org/10.1186/1471-2105-8-S4-S8
Burger L, van Nimwegen E (2010) Disentangling direct from indirect co-evolution of residues in protein alignments. PLoS Comput Biol 6:1000633. https://doi.org/10.1371/journal.pcbi.1000633
Chen XW, Liu M (2005) Prediction of protein-protein interactions using random decision forest framework. Bioinformatics 21:4394–4400. https://doi.org/10.1093/bioinformatics/bti721
van Wijk SJL, de Vries SJ, Kemmeren P et al (2009) A comprehensive framework of E2–RING E3 interactions of the human ubiquitin–proteasome system. Mol Syst Biol 5:295. https://doi.org/10.1038/msb.2009.55
Kerrien S, Aranda B, Breuza L et al (2012) The IntAct molecular interaction database in 2012. Nucleic Acids Res 40:D841–D846. https://doi.org/10.1093/nar/gkr1088
Salwinski L, Miller CS, Smith AJ et al (2004) The database of interacting proteins: 2004 update. Nucleic Acids Res 32:D449–D451. https://doi.org/10.1093/nar/gkh086
Keshava Prasad TS, Goel R, Kandasamy K et al (2009) Human protein reference database - 2009 update. Nucleic Acids Res 37:767–772. https://doi.org/10.1093/nar/gkn892
Zanzoni A, Montecchi-Palazzi L, Quondam M et al (2002) MINT: a molecular INTeraction database. FEBS Lett 513:135–140
Mewes H, Frishman D et al (2002) MIPS: a database for genomes and protein sequences. Nucleic Acids Res 30:31
Stark C, Breitkreutz B, Reguly T et al (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34:D35
Das J, Yu H (2012) HINT: high-quality protein interactomes and their applications in understanding human disease. BMC Syst Biol 6:92. https://doi.org/10.1186/1752-0509-6-92
Kuhn M, von Mering C, Campillos M et al (2008) STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res 36:D684–D688. https://doi.org/10.1093/nar/gkm795
Szklarczyk D, Morris J, Cook H et al (2017) The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 45:D362
Davis F, Sali A (2005) PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics 21:1901
Stein A, Russell R, Aloy P (2005) 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res 33:D413
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Kutmon M, van Iersel MP, Bohler A et al (2015) PathVisio 3: an extendable pathway analysis toolbox. PLoS Comput Biol 11:e1004085. https://doi.org/10.1371/journal.pcbi.1004085
Dubois J, Cottret L, Ghozlane A et al (2012) Systrip: a visual environment for the investigation of time-series data in the context of metabolic networks. IEEE, Washington, DC
Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ (2005) MAPPIS: multiple 3D alignment of protein-protein interfaces. In: Lecture notes in computer science (including subseries Lecture notes in artificial intelligence and Lecture notes in bioinformatics). Springer, Berlin, pp 91–103
Murakami Y, Mizuguchi K (2018) PSOPIA: toward more reliable protein-protein interaction prediction from sequence information. In: ICIIBMS 2017 - 2nd International Conference on Intelligent Informatics and Biomedical Sciences. Institute of Electrical and Electronics Engineers Inc, Washington, DC, pp 255–261
Prieto C, de Las RJ (2006) APID: agile protein interaction DataAnalyzer. Nucleic Acids Res 34:W298–W302. https://doi.org/10.1093/nar/gkl128
Garcia-Garcia J, Valls-Comamala V, Guney E et al (2017) iFrag: a protein–protein interface prediction server based on sequence fragments. J Mol Biol 429:382–389. https://doi.org/10.1016/j.jmb.2016.11.034
Mosca R, Céol A, Aloy P (2013) Interactome3D: adding structural details to protein networks. Nat Methods 10:47
Segura Mora S, Assi SA, Fernandez-Fuentes N (2010) Presaging critical residues in protein interfaces-web server (PCRPi-W): a web server to chart Hot spots in protein interfaces. PLoS One 5:e12352. https://doi.org/10.1371/journal.pone.0012352
Sukhwal A, Sowdhamini R (2015) PPcheck: a webserver for the quantitative analysis of protein-protein interfaces and prediction of residue hotspots. Bioinformatics Biol Insights 9:141–151. https://doi.org/10.4137/BBI.S25928
Bösl K, Ianevski A, Than TT et al (2019) Common nodes of virus–host interaction revealed through an integrated network analysis. Front Immunol 10:2186. https://doi.org/10.3389/fimmu.2019.02186
Vignuzzi M, López CB (2019) Defective viral genomes are key drivers of the virus–host interaction. Nat Microbiol 4:1075–1087
Navratil V, de Chassey B, Meyniel L et al (2009) VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res 37:661–668. https://doi.org/10.1093/nar/gkn794
Enjuanes L, Almazán F, Sola I, Zuñiga S (2006) Biochemical aspects of coronavirus replication and virus-host interaction. Annu Rev Microbiol 60:211–230. https://doi.org/10.1146/annurev.micro.60.080805.142157
Zheng YH, Jeang KT, Tokunaga K (2012) Host restriction factors in retroviral infection: promises in virus-host interaction. Retrovirology 9:112
Cheng F, Liu C, Jiang J et al (2012) Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol 8(5):e1002503
Yildirim MA, Goh KI, Cusick ME, Barabási AL, Vidal M (2007) Drug-target network. Nat Biotechnol 25(10):1119
Mestres J, Gregori-Puigjané E, Valverde S et al (2009) The topology of drug–target interaction networks: implicit dependence on drug properties and target families. Mol Biosyst 5:1051
Janga S, Tzakos T (2009) Structure and organization of drug-target networks: insights from genomic approaches for drug discovery. Mol Biosyst 5:1536
Vogt I, Mestres J (2010) Drug-target networks. Mol Inform 29:10–14. https://doi.org/10.1002/minf.200900069
Jalencas X, Mestres J (2013) On the origins of drug polypharmacology. Med Chem Commun 4:80
Boran A, Iyengar R (2010) Systems approaches to polypharmacology and drug discovery. Curr Opin Drug Discov 13:297
Peters JU (2013) Polypharmacology - foe or friend? J Med Chem 56:8955–8971
Reddy A, Zhang S (2013) Polypharmacology: drug discovery for the future. Exp Rev Clin Pharmacol 6:41–47
Awale M, Reymond JL (2017) The polypharmacology browser: a web-based multi-fingerprint target prediction tool using ChEMBL bioactivity data. J Cheminform 9:11. https://doi.org/10.1186/s13321-017-0199-x
Chen B, Wild D, Guha R (2009) PubChem as a source of polypharmacology. J Chem Inf Model 49:2044–2055. https://doi.org/10.1021/ci9001876
Berger S, Iyengar R (2009) Network analyses in systems pharmacology. Bioinformatics 25:2466
Hopkins AL (2008) Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 4:682–690
Zhang Y, Cheng X, et al 2012 Drug repositioning: an important application of network pharmacology. en.cnki.com.cn
Ye H, Wei J, Tang K, Feuers R, Hong H (2016) Drug repositioning through network pharmacology. Curr Top Med Chem 16:3646. https://doi.org/10.2174/1568026616666160530181328
von Eichborn J, Murgueitio M et al (2010) PROMISCUOUS: a database for network-based drug-repositioning. Nucleic Acids Res 39:D1060
Sydow D, Burggraaff L, Szengel A et al (2019) Advances and challenges in computational target prediction. ACS Publ 59:1728–1742. https://doi.org/10.1021/acs.jcim.8b00832
Messina F, Giombini E, Agrati C et al (2020) COVID-19: viral-host interactome analyzed by network based-approach model to study pathogenesis of SARS-CoV-2 infection. J Transl Med 18:233. https://doi.org/10.1186/s12967-020-02405-w
Hazafa A, ur-Rahman K, Haq I et al (2020) The broad-spectrum antiviral recommendations for drug discovery against COVID-19. Drug Metab Rev 52:408–424
Gordon D, Jang G, Bouhaddou M et al (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583:459
Zhou Y, Hou Y, Shen J et al (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6:14
Gysi DM, do Valle Í, Zitnik M et al (2020) Network medicine framework for identifying drug repurposing opportunities for COVID-19. ArXiv
Mu C, Sheng Y, Wang Q et al (2020) Potential compound from herbal food of rhizoma polygonati for treatment of COVID-19 analyzed by network pharmacology and molecular docking technology. J Funct Foods:104149
Sadegh S, Matschinske J, Blumenthal DB et al (2020) Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat Commun 11:3518
Dominguez Andres A, Feng Y, Rosa Campos A et al SARS-CoV-2 ORF9c is a membrane-associated protein that suppresses antiviral responses in cells. BioRxiv. https://doi.org/10.1101/2020.08.18.256776
Martin R, Löchel H, Welzel M et al (2020) CORDITE: the curated CORona Drug InTERactions database for SARS-CoV-2. iScience 23:101297
Xu J, Xu X, Jiang L et al (2020) SARS-CoV-2 induces transcriptional signatures in human lung epithelial cells that promote lung fibrosis. Respir Res 21:182. https://doi.org/10.1186/s12931-020-01445-6
Kumar S (2020) COVID-19: a drug repurposing and biomarker identification by using comprehensive gene-disease associations through protein-protein interaction network analysis. Preprints. https://doi.org/10.20944/preprints202003.0440.v1
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC
About this protocol
Cite this protocol
Kumar, S. (2021). Protein–Protein Interaction Network for the Identification of New Targets Against Novel Coronavirus. In: Roy, K. (eds) In Silico Modeling of Drugs Against Coronaviruses. Methods in Pharmacology and Toxicology. Humana, New York, NY. https://doi.org/10.1007/7653_2020_62
Download citation
DOI: https://doi.org/10.1007/7653_2020_62
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1365-8
Online ISBN: 978-1-0716-1366-5
eBook Packages: Springer Protocols