Abstract
Patient-derived or genomically modified human induced pluripotent stem cells (iPSCs) offer the opportunity to study neurodevelopmental and neurodegenerative disorders. Overexpression of certain neurogenic transcription factors (TFs) in iPSCs can induce efficient differentiation into homogeneous populations of the disease-relevant neuronal cell types. Here we provide protocols for genomic manipulations of iPSCs by CRISPR/Cas9. We also introduce two methods, based on lentiviral delivery and the piggyBac transposon system, to stably integrate neurogenic TFs into human iPSCs. Furthermore, we describe the TF-mediated neuronal differentiation and maturation in combination with astrocyte cocultures.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
- Human induced pluripotent stem cells
- Nucleofection
- PiggyBac transposon
- Lentiviral transduction
- CRISPR/Cas9
- Transcription factor-mediated neuronal differentiation
- Astrocyte coculture
1 Introduction
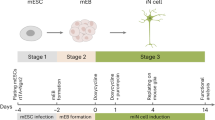
Induced pluripotent stem cells (iPSCs) enable studying neurodevelopmental and neurodegenerative diseases such as autism spectrum disorders including fragile X syndrome and Rett syndrome, amyotrophic lateral sclerosis, Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, or spinal muscular atrophy [1]. Human iPSC lines are generated by reprogramming of fibroblasts, hair, or blood samples [2], which are either directly donated by patients with a disease-relevant phenotype and a known genotype or disease-causing mutations can be introduced into the genome of the iPSCs by genomic modifications such as CRISPR/Cas9 [3]. To study the effect of the mutations on the cellular level, iPSCs can be differentiated into the disease-relevant neuronal subtypes. Conventional differentiation protocols rely on the addition of specific soluble growth factors and compounds to the culturing media. These factors trigger intracellular signaling pathways affecting transcription factors (TFs), which in turn induce neuronal differentiation by changing gene expression levels and triggering gene regulatory networks. However, these protocols can be very delicate and time-consuming, lasting from several weeks to months, and yield a heterogeneous mixture of different neuronal subtypes at different developmental stages and glia cells. The forced expression of certain neurogenic TFs in human iPSCs shortcuts neuronal differentiation resulting in rapid neurogenesis that yields highly homogeneous populations of neurons [4,5,6,7]. Here we describe the culturing of a robust inducible-neuronal iPSC line as well as different methods to introduce neurogenic TFs and genomic modifications into human iPSCs and how to differentiate those iPSCs into mature neurons.
Neurogenic TFs under the control of a doxycycline-inducible promoter can be stably integrated in the genome of iPSCs either by lentiviral delivery [8] or via the piggyBac transposon system [9]. While lentiviruses have a high efficiency in delivering transgenes, the preparation of viral particles is laborious, time-consuming and requires biosafety level 2. In contrast, the piggyBac transposon system offers a nonviral alternative to efficiently cut and paste transgenes into the genome. The production of plasmids is faster and cheaper and the piggyBac system requires only standard laboratory biosafety levels. For genome editing of human iPSCs with great precision, the CRISPR/Cas9 technology is the method of choice since it is easy-to-use, efficient, and cost-effective. Genomically modified iPSCs can be differentiated into neurons by doxycycline-induced overexpression of TFs and maturation is achieved by astrocyte coculture.
2 Materials
2.1 Human iPSC Culture
-
1.
Human iPSC line with or without genomic modifications (e.g., PGP1 cells (Personal Genome Project iPS cell line, derived from Participant #1 (PGP1, hu43860C)), can be obtained from Coriell #GM23338, the matching primary fibroblast line is #GM23248) or iNGN cells (modified from PGP1, contains the neurogenic TFs Neurogenin-1 and Neurogenin-2 under the control of the doxycycline-inducible promoter. This cell line is part of the ENCODE catalogue (https://www.encodeproject.org) #ENCBS369AAA [4]).
-
2.
Coating medium: DMEM medium, 1% penicillin–streptomycin. Store at 4 °C.
-
3.
Matrigel hESC-qualified matrix (Corning). Store aliquots at −20 °C (see Note 1 ).
-
4.
mTeSR™1 medium: 400 ml mTeSR™1 Basal medium (Stemcell Technologies), 100 ml mTeSR™1 5× Supplement (Stemcell Technologies), 5 ml penicillin–streptomycin, sterile-filter (0.45 μm). Store at 4 °C for a maximum of 2 weeks. Do not prewarm before usage.
-
5.
mTeSR™1 medium with ROCK inhibitor (ROCKi): add ROCK inhibitor to mTeSR™1 medium (3.3 μg/ml final concentration). Store at 4 °C for a maximum of 2 weeks.
-
6.
Dissociation reagent: TrypLE Express (Thermo Fisher Scientific). Store at room temperature.
-
7.
Freezing medium: mFreSR™ (Stemcell Technologies). Store aliquots at −20 °C.
-
8.
1× PBS pH 7.2 without calcium and magnesium. Store at room temperature.
2.2 Nucleofection of PiggyBac Plasmids
-
1.
4D-Nucleofector™ System: 4D-Nucleofector™ Core Unit and 4D-Nucleofector™ X-Unit (Lonza).
-
2.
P3 Primary Cell 4D-Nucleofector™ X Kit: Containing the Nucleofector™ Solution, Supplement and 100 μl Cuvettes (Lonza). Store the Nucleofector™ Solution and the Supplement at 4 °C.
-
3.
PiggyBac vector containing the gene of interest under the control of a doxycycline-inducible promoter, such as Addgene plasmid #104454 (see Note 2 and Fig. 2a). Store at −20 °C.
-
4.
Transposase vector, such as System Biosciences #PB210PA-1. Store at −20 °C.
-
5.
Antibiotic: If you would like to select the cells for the integrated piggyBac construct, use the appropriate antibiotic (e.g., blasticidin or puromycin). Store aliquots at −20 °C. After thawing store at 4 °C, protected from light.
-
6.
PiggyBac copy number kit (System Biosciences #PBC100A-1) including UCR1 primer mix, PBcopy primer mix, and cell lysis buffer. Store at 4 °C.
-
7.
Real time PCR master mix, such as Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific). Store at 4 °C.
-
8.
Real time PCR system, such as StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific).
2.3 Lentivirus Production and Transduction
-
1.
A highly transfectable cell line, such as 293T/17 cells (a gift from Didier Trono; ATCC® #CRL-11268).
-
2.
DMEM with 10% FBS: DMEM medium, 10% FBS, 1% penicillin–streptomycin. Store at 4 °C.
-
3.
DMEM w/o FBS: DMEM medium, 1% penicillin–streptomycin. Store at 4 °C.
-
4.
1 mg/ml PEI solution (pH 7.1): adjust the pH with 0.1 N NaOH, sterile-filter (0.22 μm), aliquot, and store at −20 °C. After thawing, working aliquots can be stored at 4 °C.
-
5.
Lentiviral vector containing the gene of interest under a doxycycline-inducible promoter (e.g., pLV_TRET_Ngn2-2A-Ngn1 (Addgene plasmid #61471) [4] and pLV_hEF1a_rtTA3 (Addgene plasmid #61472) [4] or pLIX403 (Addgene plasmid #41395)) (see Note 3 and Fig. 3a, b). Store at −20 °C.
-
6.
Viral packaging plasmid psPAX2 (a gift from Didier Trono, Addgene plasmid #12260). Store at −20 °C.
-
7.
Viral envelope plasmid pMD2G (a gift from Didier Trono, Addgene plasmid #12259). Store at −20 °C.
-
8.
50% PEG 6000 solution. Store at 4 °C.
-
9.
4 M NaCl solution. Store at 4 °C.
-
10.
PBS pH 7.2 without calcium and magnesium. Store at room temperature.
-
11.
Lenti-X™ GoStix™ (Clontech).
-
12.
Antibiotic: If you would like to select the cells for the integrated lentiviral construct, use the appropriate antibiotic (such as blasticidin or puromycin). Store aliquots at −20 °C, after thawing store at 4 °C, protected from light.
-
13.
DNA extraction kit (e.g., DNeasy® Blood and Tissue Kit (Qiagen)).
-
14.
Albumin plasmid pAlbumin (a gift from Didier Trono, Addgene plasmid #22037). Store at −20 °C.
-
15.
TaqMan® PCR master mix, such as TaqMan® Universal PCR Master Mix (Thermo Fisher Scientific). Store at 4 °C.
-
16.
TaqMan® primer and probes for WPRE and albumin detection (see Table 1). Dilute in ddH2O to a concentration of 10 μM and store at −20 °C.
-
17.
Real time PCR system, such as StepOnePlus™Real-Time PCR System (Thermo Fisher Scientific).
2.4 Genome Editing with CRISPR/Cas9
-
1.
Cas9-sgRNA construct , such as pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene plasmid #62988) or pSpCas9(BB)-2A-GFP (PX458) (Addgene plasmid #48138), which is expressing both sgRNA together with the gRNA scaffold as well as the staphylococcus pyogenes Cas9 [10]. Store at −20 °C.
-
2.
10× T4 Ligation Buffer (New England Biolabs). Store at −20 °C.
-
3.
T4 Polynucleotide Kinase (New England Biolabs). Store at −20 °C.
-
4.
BbsI restriction enzyme (10 U/μl) (Thermo Fisher Scientific). Store at −20 °C.
-
5.
Buffer G (Thermo Fisher Scientific). Store at −20 °C.
-
6.
Calf Intestinal Alkaline Phosphatase (CIP) (New England Biolabs). Store at −20 °C.
-
7.
Electrophoresis gel and chamber.
-
8.
Gel extraction kit, such as QIAquick® Gel Extraction Kit (Qiagen).
-
9.
Ligation kit, such as Mighty Mix ligation kit (Clontech). Store at −20 °C.
-
10.
Chemically competent bacteria (e.g., Stbl3). Store at −80 °C.
-
11.
S.O.C. medium. Store at 4 °C.
-
12.
LB-Antibiotics plates with 100 μg/ml ampicillin. Store at 4 °C.
-
13.
Miniprep kit, such as QIAprep® Spin Miniprep Kit (Qiagen).
-
14.
Sequencing primer , such as 5′-TTTCTTGGGTAGTTTGCAGTTTT-3′. Dilute in ddH2O to a concentration of 10 μM and store at −20 °C.
-
15.
4D-Nucleofector™ System (see Subheading 2.2).
2.5 Doxycycline-Induced Differentiation
-
1.
Poly-l-lysine (PLL) solution: Dilute PLL hydrobromide in ddH2O to a stock concentration of 1 mg/ml. Store at 4 °C.
-
2.
Laminin solution: 1 mg/ml stock. Store aliquots at −20 °C.
-
3.
1× PBS with calcium and magnesium. Store at 4 °C.
-
4.
Doxycycline solution: dissolve 10 mg doxycycline hyclate powder in 20 ml PBS (0.5 mg/ml = 1000×), sterile-filter (0.22 μm). Store aliquots at −20 °C; after thawing store at 4 °C, protected from light.
-
5.
Differentiation medium: mTeSR™1 medium supplemented with 0.5 μg/ml doxycycline. Store at 4 °C for a maximum of 2 weeks.
-
6.
Maturation medium: 10 ml BrainPhys™ Neuronal Medium (Stemcell Technologies) supplemented with 200 μl NeuroCult™ SM1 Neuronal Supplement (Stemcell Technologies), 100 μl N2 Supplement-A (Stemcell Technologies), 20 μl of 10 μg/ml recombinant Human BDNF to a final concentration of 20 ng/ml (Peprotech), 20 μl of 10 μg/ml recombinant Human GDNF to a final concentration of 20 ng/ml (Peprotech), 98 μl of 50 mg/ml dibutyryl cAMP to a final concentration of 1 mM (Sigma), 50 μl of 40 mM ascorbic acid to a final concentration of 200 nM (Sigma), and 100 μl of 100 × penicillin–streptomycin (see Note 4 ). Mix thoroughly. Store at 4 °C for a maximum of 2 weeks.
2.6 Coculturing with Astrocytes
-
1.
Rat primary cortical astrocytes (Thermo Fisher Scientific).
-
2.
Astrocyte medium: DMEM +4.5 g/l d-Glucose +1 mM Pyruvate supplemented with N2 Supplement (Thermo Fisher Scientific), 10% OneShot fetal bovine serum (Thermo Fisher Scientific) and 1% penicillin–streptomycin. Store aliquots at −20 °C. After thawing, working aliquots can be stored at 4 °C for a maximum of 2 weeks.
-
3.
Accutase. Store aliquots at −20 °C.
-
4.
1 M HCl. Store at room temperature.
-
5.
100% ethanol. Store at room temperature.
-
6.
Low-melting paraffin. Store at room temperature.
-
7.
1× PBS with calcium and magnesium. Store at 4 °C.
-
8.
Ara-C: dissolve 11 mg cytosine β-d-arabinofuranoside hydrochloride powder in 15 ml ddH2O (2.5 mM = 500×), sterile-filter (0.22 μm). Store aliquots at −20 °C, after thawing store at 4 °C for several weeks, protected from light.
-
9.
1% BSA: 8.7 ml BrainPhys™ Neuronal Medium (Stemcell Technologies) supplemented with 128 μl 1 M HEPES and 1.33 μl 7.5% BSA. Prewarm and use immediately.
-
10.
0.2% BSA: 4 ml BrainPhys™ Neuronal Medium (Stemcell Technologies) supplemented with 59 μl 1 M HEPES and 1 ml 1% BSA from previous step. Prewarm and use immediately.
-
11.
Minimal maturation medium (see Note 5 ): 10 ml BrainPhys™ Neuronal Medium (Stemcell Technologies) supplemented with 200 μl NeuroCult™ SM1 Neuronal Supplement (Stemcell Technologies), 100 μl N2 Supplement-A (Stemcell Technologies), 50 μl of 40 mM ascorbic acid (dissolved in ddH2O) to a final concentration of 200 nM (Sigma) and 100 μl of 100 × penicillin–streptomycin (see Note 4 ). Mix thoroughly. Store at 4 °C for a maximum of 2 weeks.
3 Methods
3.1 Human iPSC Culture
-
1.
For the coating of cell culture plates, resuspend one aliquot of Matrigel in the appropriate amount of cold coating medium (see Note 1 ). Add the Matrigel solution to the cell culture plates and distribute equally so that the entire well is covered (see Note 6 ). Incubate at room temperature for at least 45 min. Use immediately or store at 4 °C for a maximum of 2 weeks. Prior to use, simply aspirate the coating medium and add the cell suspension. No washing step is required.
-
2.
To thaw iPSCs, get the frozen vials from the liquid nitrogen tank and keep on dry ice. Thaw carefully in a 37 °C water bath or alternatively with the ThawSTAR™ Automated Cell Thawing System (BioCision™) until only a small ice cube remains. Transfer the cell solution to a 15 ml Falcon tube and add dropwise 2–3 ml 1× PBS w/o Ca2+ and Mg2+. Spin down at 400 × g for 4 min and aspirate the supernatant. Resuspend the cell pellet in mTeSR™1 with ROCKi and transfer to a cell culture plate coated with Matrigel and place in the incubator (37 °C, 5% CO2). After 24 h, wash the cells once with 1× PBS w/o Ca2+ and Mg2+ and change the medium to mTeSR™1 w/o ROCKi. Change the medium every day until the next passaging (see Note 7 ).
-
3.
In order to passage iPSCs, aspirate the culture medium and wash the cells once with 1× PBS w/o Ca2+ and Mg2+. Dissociate the cells by adding TrypLE and place in the incubator for approximately 2–3 min. Add 1× PBS w/o Ca2+ and Mg2+ and pipet up and down to collect all cells. Transfer the cell solution to a 15 ml Falcon tube and spin down at 400 × g for 4 min. Aspirate the supernatant and resuspend the cell pellet in mTeSR™1 with ROCKi. Count the cells using Trypan Blue (e.g., with the Countess™ II FL Automated Cell Counter or hemocytometer) and seed the appropriate number of cells in Matrigel- or poly-l-lysine + laminin-coated cell culture plates (see Note 8 ). Mix well and place in the incubator (37 °C, 5% CO2). After 24 h, wash the cells once with 1× PBS w/o Ca2+ and Mg2+ and change the medium to mTeSR™1 w/o ROCKi. Change the medium every day until next passaging (see Note 7 and Fig. 1). It is recommended to check the iPSCs regularly for mycoplasma contamination (see Note 9 ).
-
4.
To freeze iPSCs, dissociate the cells with TrypLE and collect in 1× PBS w/o Ca2+ and Mg2+, spin down and resuspend the pellet in mFreSR™ medium. If necessary, count the cells and aliquot the appropriate amount into cryotubes (see Note 10 ). Put the tubes in a freezing container and store at −80 °C for at least 2 h. Subsequently, store in liquid nitrogen.
Representative images of human iPSCs [4] grown on Matrigel-coated cell culture plates. (a) iPSCs 1 day after passaging in mTeSR™1 with ROCKi. (b) iPSC colonies in mTeSR™1 w/o ROCKi. (c) Confluent iPSC colonies. Scale bar represents 200 μm
3.2 Nucleofection of iPSCs
-
1.
In order to electroporate piggyBac and transposase vectors into iPSCs in suspension, use the X-Unit of the 4D-Nucleofector™ System in combination with the P3 Primary Cell 4D-Nucleofector™ X Kit according to the manufacturer’s guidelines.
-
2.
First of all, prepare the DNA, the Nucleofector™ solution and the cell culture plates. For a nucleofection reaction in 100 μl cuvettes, mix 10 μg piggyBac vector and 2.5 μg transposase vector in less than 10 μl volume (maximum 10% of the final sample volume) in a 1.5 ml tube. In a separate tube, mix 82 μl Nucleofector™ solution with 18 μl supplement per nucleofection reaction and bring to room temperature. Prepare Matrigel-coated cell culture plates with the desired volume of mTeSR™1 medium with ROCKi and prewarm in the incubator (see Note 11 ).
-
3.
Switch on the X-Unit of the 4D-Nucleofector™ System and choose the cell-type specific program for the human embryonic stem cell line H9, the cuvette size, P3 primary solution and the pulse CB-156 or CB-150 (see Note 12 ).
-
4.
Dissociate the cells to be nucleofected using TrypLE, centrifuge (400 × g, 4 min) and resuspend in mTeSR™1 with ROCKi. Determine the cell number, transfer 800,000 cells for each nucleofection into a 1.5 ml tube and centrifuge (400 × g, 4 min). Aspirate the supernatant and resuspend the cells in 100 μl room temperature Nucleofector™ solution with supplement, mix with the DNA and transfer into an electroporation cuvette and close the lid. Avoid air bubbles while pipetting. Gently tap the cuvette to make sure that the sample covers the bottom.
-
5.
Quickly put the cuvette(s) into the Nucleofector™ and press the start button to apply the pulse CB-156 or CB-150. Immediately after, carefully remove the samples, add mTeSR™1 with ROCKi into the cuvette, mix by gently pipetting up and down two to three times and transfer the complete solution onto a Matrigel-coated plate with prewarmed medium and place in the incubator (see Note 13 ).
-
6.
The next day, wash the cells with 1× PBS w/o Ca2+ and Mg2+ and change the medium to mTeSR™1 w/o ROCKi. Change the medium every day until next passaging (see Fig. 2b). Starting 48 h after nucleofection, select the cells with an integrated construct with the appropriate antibiotic (see Note 14 ).
-
7.
In order to determine the number of the integrated piggyBac constructs, use the piggyBac copy number kit from System Biosciences (see Note 15 ). To prepare genomic DNA, seed the cells in a 12-well plate (see Note 16 ). When confluent, wash once with 1× PBS w/o Ca2+ and Mg2+ and add 250 μl lysis buffer to each well. Freeze the cells at −80 °C and thaw the plate at room temperature to ensure complete cellular lysis. Detach the cells by pipetting up and down, transfer the lysates to 1.5 ml tubes and heat them at 95 °C for 2 min. Centrifuge at 17,000 × g for 2 min and transfer the supernatant to a new 1.5 ml tube. The lysates should be placed on ice if used immediately or stored at −20 °C.
-
8.
Prepare two master mixes of 4.75 μl ddH2O, 6.25 μl SYBR® Green, and 0.5 μl primers per sample (one master mix with UCR1 primers for genomic DNA detection and one with PBcopy primers for piggyBac detection). Aliquot 12 μl master mix per well of a 96-well plate and add 0.5 μl lysate (≤500 ng DNA). Seal the plate, carefully mix by vortexing and briefly spin down. Run the qPCR with the following program: 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 95 °C for 15 s and 60 °C for 1 min, followed by 15 s at 95 °C, 15 s at 60 °C, and 15 s at 95 °C (see Note 17 ).
-
9.
Calculate the copy number as follows [11]: ΔΔCt = 2(average PBcopy Ct–average UCR1 Ct), divide the ΔΔCt by 2 as there are two copies of the UCR1 sequence per genome.
Nucleofection of iPSCs with the piggyBac plasmid. (a) Schematic representation of the piggyBac plasmid containing the 5′ and 3′ inverted terminal repeats (ITR), core insulator (Core I), the doxycycline-inducible TRE promoter driving the expression of a transcription factor (TF) or in our example of EGFP which can be excised be the restriction enzymes NheI and XhoI and replaced by a TF of interest, followed by a V5 tag and a bGH poly A signal. Furthermore, the plasmid contains an EF1α promoter driving the expression of the doxycycline-sensitive transactivator rtTA followed by a T2A signal and a puromycin resistance gene. (b) Representative images of iPSCs nucleofected with the plasmid depicted in (a) 24 and 48 h after doxycycline (Dox) induction and respective controls without doxycycline (no Dox CTRL). Scale bar represents 200 μm
3.3 Lentivirus Production and Transduction
-
1.
For the production and transduction of lentiviruses, titration and copy number determination, we follow the protocol from the Trono lab [8].
-
2.
One day prior to transfection, seed 8,000,000,293T/17 cells in a 10 cm culture dish. The next day, replace the culture medium with 4 ml fresh DMEM with 10% FBS. The cells are transfected using 45 μg of polyethylenimine (PEI) combined with 15 μg DNA containing the plasmid of interest (see Fig. 3a, b), the viral packaging (psPAX2) plasmid, and the viral envelope (pMD2G) plasmid in a 4:2:1 ratio. In detail, mix 45 μl of 1 mg/ml PEI solution with 955 μl DMEM medium w/o FBS in a 1.5 ml tube. In a separate tube, mix 2.1 μg pMD2G, 4.2 μg psPAX2, and 8.4 μg vector of interest with 1 ml DMEM w/o FBS. Combine PEI and DNA mix and incubate at room temperature for 15–30 min. Subsequently, add the transfection mix dropwise to the 293T/17 cells and place the culture dish into the incubator (see Note 18 ).
-
3.
The next day, replace the medium with 7 ml DMEM with 10% FBS. After 24 h, collect the supernatant, with the help of a syringe pass it through a 0.45 μm PES filter into a 50 ml Falcon tube and store at 4 °C. Add 7 ml fresh DMEM with 10% FBS and after another 24 h collect the supernatant, filter and pool with the first collection.
-
4.
Mix 14 ml of cell culture supernatant (48 h and 72 h harvests) with 6.6 ml of diluted polyethyleneglycol (PEG) solution containing 3.5 ml of 50% PEG 6000 solution, 1.5 ml of 4 M NaCl solution, and 1.6 ml of 1× PBS w/o Ca2+ and Mg2+, and keep at 4 °C overnight or over the weekend. Centrifuge the tubes at 7000 g for 10 min at 4 °C (see Note 19 ). After the centrifugation, a white pellet should be visible. Carefully decant the supernatant and resuspend the pellet in 150 μl 1× PBS w/o Ca2+ and Mg2+ by pipetting up and down. Vigorously vortex the tubes for 20–30 s to further resuspend the pellets. Lenti-X™ GoStix™ can be used to confirm the successful generation of viral particles. Transfer the virus suspension into 1.5 ml screw-cap tubes in aliquots of 50 μl, snap-freeze in crushed dry ice and store at −80 °C.
-
5.
In order to transfect iPSCs with the viral particles, the cells should be approximately 40 ± 10% confluent. Wash the cells once with 1× PBS w/o Ca2+ and Mg2+ and add mTeSR™1 medium to the well. Add mTeSR™1 to the tube containing the virus suspension, mix gently and transfer dropwise to one or more wells (see Note 20 ). Incubate the culture dish overnight and change the medium on the next day (see Note 21 ). Starting 48 h after transduction, select for cells with an integrated construct with the appropriate antibiotic (see Note 14 ).
-
6.
For titration and copy number determination, we perform a qPCR on genomic DNA to count the number of integrated viral particles (WPRE) relative to the genome (albumin gene). Wait at least 96 h before isolating genomic DNA using a genomic DNA extraction kit, such as the DNeasy® Blood and Tissue Kit (Qiagen), according to the manufacturer’s instructions. The DNA should be placed on ice if used immediately or stored at −20 °C.
-
7.
Adjust the concentration of the pAlbumin plasmid used for normalization to 1 mg/ml, which corresponds to 1.2 × 1011 molecules/μl (see Note 22 ). Prepare a standard curve, with the first point being 1 × 107 molecules in 8 μl (which corresponds to 1.25 × 106 molecules/μl). Prepare the other points of the standard curve by serial tenfold dilutions until there are ten molecules in 8 μl (see Fig. 3c).
-
8.
Prepare two master mixes of 8.5 μl TaqMan® Universal PCR Master Mix, 0.17 μl forward primer (10 μM), 0.17 μl reverse primer (10 μM), and 0.17 μl probe (10 μM) per sample including all samples and standards in duplicates (one master mix with albumin primers and probe for genomic DNA detection and one with WPRE primers and probe for lentivirus detection). Aliquot 9 μl master mix per well of a 96-well plate and add 8 μl of sample DNA (concentration between 50 and 100 ng DNA in 8 μl). Seal the plate, carefully mix by vortexing and briefly spin down. Run the qPCR with the settings for FAM fluorochromes and BHQ quenchers with the following program: 1 cycle of 10 min at 95 °C followed by 50 cycles of 15 s at 95 °C and 1 min at 60 °C (see Note 17 ).
-
9.
Plot the standard curve using the software of your qPCR machine or manually using other software such as Microsoft Excel, and calculate the quantity of albumin and WPRE for each sample using the equation of the standard curve.
-
10.
Calculate the copy number for each sample as follows: Copy number = (quantity mean of WPRE sequence/quantity mean of Alb sequence) × 2.
-
11.
Calculate the viral titer with the following formula: Titer (viral genome/ml) = (number of target cells counted at day 1 × number of copies per cell of the sample)/volume of supernatant (ml).
Transduction of iPSCs with lentiviral vectors. (a) and (b) Schematic representation of the lentiviral constructs. (a) The pLV system consists of two constructs: pLV_hEF1a_rtTA3 (top) expresses the rtTA transactivator under the control of the constitutively active EF1α promoter and pLV_TRET_Ngn2-2A-Ngn1 (bottom) expresses the transgenes under the control of the doxycycline-inducible TRE promotor [4]. The pLV plasmids depicted to here do not contain any selection markers. If selection is required, antibiotic resistance genes should be cloned into the plasmids. (b) The pLIX403 system is an “all-in-one” doxycycline-inducible system. It expresses a puromycin resistance gene together with the rtTA transactivator under the control of the constitutively active PGK promotor and the transgene under the control of the doxycycline-inducible TRE promotor. 5′ and 3′ LTR—long terminal repeats, WPRE—woodchuck hepatitis virus posttranscriptional regulatory element. (c) Schematic representation of the serial dilution of the pAlbumin plasmid for titration and copy number determination (modified from [8])
3.4 Genome Editing with CRISPR/Cas9
-
1.
Design a sgRNA for the locus of interest using designing tools (such as http://crispr.mit.edu/, see Note 23 ) [12].
-
2.
Order two oligos representing the sgRNA with a design as follows: top oligo—CACC(G)[20 N of sgRNA], bottom oligo—AAAC[20 N reverse complement of sgRNA](C) [10] (see Note 24 ).
-
3.
Anneal the oligos in a thermocycler using 2 μl top oligo, 2 μl bottom oligo, 2 μl 10× T4 Ligation Buffer, T4 Polynucleotide Kinase, and 13 μl ddH2O and program the thermocycler with 30 min at 37 °C, 5 min at 95 °C, a ramp down to 25 °C (ramp rate of 5 °C/min) and hold at 4 °C. The oligo hybrid can directly be used for cloning into the linearized PX459 or PX458 vector.
-
4.
Cut and dephosphorylate the vector using 2 μl BbsI with 3 μg of vector and fill up to 50 μl reaction volume with ddH2O (minimum of 0.5 μl BbsI for 5 μg of vector). Incubate for 1 h at 37 °C and optionally heat-inactivate the digestion for 15 min at 65 °C. Gel-purify the vector by running on an agarose gel and extracting the band using a gel extraction kit, such as QIAquick® Gel Extraction Kit (Qiagen), according to the manufacturer’s instructions. The vector can be stored at −20 °C for several months.
-
5.
Ligate sgRNA insert and vector in a 10 μl reaction with 100 ng vector, 2 μl of the oligo hybrid (1:250 diluted), 5 μl Mighty Mix, and ddH2O. Incubate the mixture for 30 min at 16 °C.
-
6.
Transform 5 μl of the reaction product into chemically competent bacteria, such as Stbl3. The rest of the reaction can be stored for up to 2 weeks. Thaw the bacteria for ~10 min on ice, add the ligation reaction, stir carefully and incubate on ice for 5–10 min. Perform a heat shock for 45 s at 42 °C transfer the sample back to ice for 2–5 min. Add 300 μl S.O.C. medium and shake the bacteria for 1 h at 37 °C and 300 rpm. Plate the bacteria on LB-plates with 100 μg/ml ampicillin and incubate overnight at 37 °C and proceed with the plasmid preparation using a Miniprep kit such as the QIAprep® Spin Miniprep Kit (Qiagen) according to the manufacturer’s guidelines.
-
7.
Check the construct for correctness using the respective sequencing primer and expand the DNA to a high concentration stock. Optionally: Test the cutting efficiency with a T7 endonuclease assay [13] (see Note 25 ).
-
8.
If homologous recombination of a disease correction or knockin of a reporter is the aim of the gene editing, a donor construct has to be provided in addition to the Cas9-sgRNA vector(s). The tag or the gene for the knockin should be framed by regions of the locus in approximately the same length as the part that is transferred into the gene (such as ~1000 bp for incorporation of an antibiotic resistance with promoter) and provided as a linearized DNA fragment. Alternatively, 5–25 bp homology can be used with microhomology-mediated end-joining [14].
-
9.
Electroporate 10 μg of the vector encoding for the correct sgRNA and optionally ~5 μg of donor construct into iPSCs (see Subheadings 2.2 and 3.2). Seed the cells into two wells of a 6-well plate.
-
10.
Using the PX459 vector, it is advisable to use 0.5–0.8 μg/ml puromycin for 12–16 h for the next night after electroporation in combination with ROCKi.
-
11.
Keep ROCKi for 2 days and check if you obtained single cells that grow to small colonies. Once the colonies are visible by eye, check half of a colony for integration or knockout of the target gene by (colony) PCR with primers binding around or in the locus that you tested for specificity before (using control DNA). Transfer the other part of each colony not used for PCR to a new well (of a 48-well plate). These are the monoclonal knockout lines usable if the PCR shows the respective shifts.
-
12.
Once a potentially positive clone has been detected, sequence the locus using Sanger sequencing, optimally with subcloning of the PCR product into a carrier vector (for instance using TOPO cloning).
3.5 Doxycycline-Induced Differentiation
-
1.
Neurons can be grown on Matrigel-coated cell culture dishes, however, especially for long-term neuronal differentiation, it is recommended to grow the neurons on cell culture plates coated with poly-l-lysine (PLL) and laminin. Dilute the PLL in ddH2O to a final concentration of 40 μg/ml, add the diluted PLL solution to the cell culture plates and distribute equally so that the entire well is covered. Incubate at 37 °C overnight and wash three times with ddH2O on the next day. Dilute the laminin in 1× PBS with Ca2+ and Mg2+ to a final concentration of 20 μg/ml and add to the PLL-coated cell culture plates. Incubate at 37 °C for approximately 4 h. Prior to use, simply aspirate the coating solution and seed the cells without washing the plates.
-
2.
Seed the iPSCs at a density of 30,000–50,000 cells per cm2 in mTeSR™1 medium with ROCKi supplemented with 0.5 μg/ml doxycycline. On the next day, wash the cells with 1× PBS w/o Ca2+ and Mg2+ and change the medium to mTeSR™1 w/o ROCKi supplemented with 0.5 μg/ml doxycycline. Change the medium daily until day 4 (Fig. 4).
-
3.
When culturing the neurons for longer time periods, it is recommended to change the stem cell medium (mTeSR™1) to maturation medium (BrainPhys™ with supplements). Change half of the medium on day 5 of differentiation to BrainPhys™ medium with supplements. Repeat changing half of the medium 2 days later. After those two adaptation medium changes, it is sufficient to change half of the medium once per week. Volume loss due to evaporation should be compensated with ddH2O.
Representative images of neuronal differentiation of human iPSCs expressing the neurogenic TFs Neurogenin-1 and Neurogenin-2 (iNGN cells) under the control of a doxycycline-inducible promoter [4]. Scale bar represents 100 μm
3.6 Coculturing with Astrocytes
-
1.
In order to increase the maturation of neurons for electrophysiological measurements, coculturing with astrocytes is highly recommended [4, 15]. We adapted the protocol from Kaech and Banker [16] to our cell culture.
-
2.
Rat primary cortical astrocytes are cultured in astrocyte medium at 37 °C and 5% CO2 according to the manufacturer’s instructions. For passaging, aspirate the culture medium and store it in a Falcon tube as a washing solution (see Note 26 ). Rinse the cells once with 1× PBS w/o Ca2+ and Mg2+. Add prewarmed Accutase and incubate the cells at 37 °C until all of them are detached (usually 5 min are sufficient). Stepwise add the cell culture medium stored in the first step to flush cells and collect all cells to a prerinsed 15 ml Falcon tube. Centrifuge at 400 × g for 5 min. Aspirate the supernatant and resuspend the pellet in prewarmed astrocyte growth medium. Count the cells using Trypan Blue and seed the appropriate amount in uncoated tissue-culture treated dishes at a seeding density of approximately 5000 cells per cm2. Change the growth medium every 3–4 days.
-
3.
For the coculture with neurons, prepare astrocytes to be ~80% confluent at day 4 of neuronal differentiation. One day before the reseeding of neurons, wash the astrocytes three times with 1× PBS w/o Ca2+ and Mg2+ and add BrainPhys™ medium with minimal supplements.
-
4.
Thoroughly clean the coverslips in a big glass petri dish. First, rinse the coverslips in ddH2O for 2 h and then shake in 50 ml 1 M HCl overnight. Rinse three times with ddH2O by shaking for 2 min, and rinse another two times with ddH2O by shaking for 2 h. Shake three times in 100% ethanol for 2 min and one time overnight. Sterilize the coverslips at 225 °C for 6–16 h (can be done overnight) (see Note 27 ) [16].
-
5.
Autoclave ~100 ml low melting paraffin in a 500 ml bottle. Melt in a boiling water bath (1 l beaker with 400 ml H2O on a heat plate with 350 °C). Use a 2 ml aspiration pipette with a 200 μl pipette tip attached and soak it in melted paraffin. Shake off extra drops and place small drops on coverslips to create paraffin feet as spacer (see Note 28 ). Coat the coverslips with PLL and laminin (see Subheading 3.5) on the side with the paraffin dots.
-
6.
Differentiate the iPSCs on Matrigel-coated cell culture dishes using 0.5 μg/ml doxycycline in mTeSR™1 medium for 4 days (see Subheading 3.5). Add 5 μM Ara-C to the culture for 1 day to remove occasionally undifferentiated cells.
-
7.
At day 5, reseed predifferentiated neurons on PLL and laminin coated coverslips with paraffin feet. Collect the old medium from the culture well and wash the cells very carefully with prewarmed 1× PBS w/o Ca2+, Mg2+. Dissociate the cells by adding Accutase and place in the incubator for approximately 5 min until the neuronal network detaches. Add the cell culture medium stored in the first step and transfer the cell solution to a 15 ml Falcon tube. Rinse the well 1–2 times with 1% BSA to collect all cells and centrifuge at 400 × g for 5 min. Aspirate the supernatant and resuspend the cell pellet in 200 μl 0.2% BSA slowly and carefully (this step is crucial for the survival of single cells). Add 800 μl of 0.2% BSA to a total volume of 1 ml. Centrifuge at 20 × g for 1 min and collect 800 μl supernatant in a fresh tube (this is the single cell suspension). Repeat the dissociation of the pellet for a maximum of five times until no pellet is visible any more. Centrifuge the single cell suspension at 400 × g for 5 min and resuspend the pellet in 0.5 ml BrainPhys™ medium. Count the cells if necessary and seed on coated coverslips on the side with the paraffin feet (see Note 29 ).
-
8.
After 2 h, place the coverslips with the differentiated iPSCs upside down into culture wells containing 80% confluent rat astrocytes. Every 7 days, exchange 50% of the BrainPhys™ medium and compensate the volume loss due to evaporation with ddH2O (see Note 30 ).
4 Notes
-
1.
Aliquot the Matrigel according to the protocol and the dilution factor provided with it (varies for each bottle of Matrigel). We prepare aliquots for dilution in 12 ml coating medium. Briefly, thaw the Matrigel on ice in the cold room or the fridge and prepare a box with dry ice to precool 1.5 ml tubes. Quickly distribute the Matrigel solution into the tubes and store at −20 °C.
-
2.
The piggyBac vector backbone can be obtained from Addgene (to be submitted, containing the EGFP gene under the control of the doxycycline-inducible promoter). For cloning of transcription factors, the EGFP can be excised using NheI and XhoI, the transcription factor cDNA can be amplified by PCR and introduced into the piggyBac vector using Gibson Assembly cloning [17].
-
3.
There are two different lentiviral vector systems that can be used: the pLV system that consists of two constructs, one expressing the rtTA transactivator from the constitutively active EF1α promoter and the other one expressing the transgene under the control of the doxycycline-inducible TRE promoter [4] (Addgene plasmids #61472 and #61471, respectively) or the pLIX403 system that expresses the rtTA transactivator and the transgene under the TRE promoter on a single construct (Addgene plasmid #41395). The pLV plasmids that are referred to in this protocol do not contain any selection markers. If selection for the integrated constructs is required, it should be cloned into the plasmids before the production of lentiviral particles.
-
4.
For better attachment of the neurons, freshly add 1 μl of a 1 mg/ml laminin solution per 1 ml supplemented BrainPhys™ medium to a final concentration of 1 μg/ml.
-
5.
For the coculture with astrocytes, we use BrainPhys™ medium with a minimal supplementation since we found that astrocytes do not grow well in the presence of cAMP. Addition of BDNF and GDNF was neither found to enhance maturation nor affect astrocytes but might be beneficial depending on experiment design.
-
6.
Use 1 ml of diluted Matrigel solution per well of a 6-well plate, 0.5 ml per well of a 12-well plate and 0.25 ml per well of a 24-well plate.
-
7.
Use 2 ml mTeSR™1 medium per well of a 6-well plate, 1 ml per well of a 12-well plate and 0.5 ml per well of a 24-well plate. If you would like to avoid feeding the cells on the weekend, add at least the 1.5-fold amount of medium on Friday.
-
8.
The optimal cell density depends on the growth rate of your iPSC line. For our cells, we seed 15,000–25,000 cells/cm2 for maintenance of stem cells, and 30,000–50,000 cells/cm2 for differentiation experiments.
-
9.
Check iPSCs in 4-week intervals for mycoplasma contamination using the Universal Mycoplasma Detection Kit (ATCC® 30-1012K™) according to the manufacturer’s instructions.
-
10.
The optimal density for freezing depends on your iPSC line. For our cells, a density of 500,000–1,000,000 cells/cryotube in 0.5–1 ml mFreSR™ works well.
-
11.
Cells of one 100 μl nucleofection reaction can be seeded to one well of a 6-well plate or distributed to multiple wells of a 12- or 24-well plate.
-
12.
The pulse CB-156 is recommended if higher transfection efficiency is favored at expenses of a lower survival rate, whereas the pulse CB-150 results in higher viability with lower transfection efficiency.
-
13.
Leaving the cells in Nucleofector™ solution for extended periods of time may lead to reduced transfection efficiency and viability so it is important to work as quickly as possible. If you face problems such as low transfection efficiency due to very big plasmids etc. you can try to incubate the cells after nucleofection in the Nucleofector™ solution at room temperature for approximately 10 min.
-
14.
The concentration of antibiotic optimal for selection depends on the specific iPSC line of choice and should be determined with a killing curve. We use a final concentration of 20 μg/ml for blasticidin, 3 μg/ml for puromycin, and 250 μg/ml for hygromycin.
-
15.
Alternatively, the copy number can be determined as described for the lentiviral transduction (see Subheading 3.3) by performing a TaqMan®-based qPCR on genomic DNA. Use the albumin gene for normalization and a gene specific for the piggyBac construct for counting the integration events. We recommend using primers and probes for the antibiotic resistance gene, if not otherwise present in the genome of your iPSC line. It is important to have both genes present on the same plasmid used for the standard curve since the preparation of the serial dilutions is prone to small variations.
-
16.
Before performing the copy number determination, the cells must be passaged at least once to avoid the interference of nonintegrated piggyBac plasmids with the qPCR.
-
17.
The optimal settings of the qPCR protocol may vary with the qPCR machine and the SYBR® Green or TaqMan® mix used. The settings described in this protocol are referring to the StepOnePlus™Real-Time PCR System (Thermo Fisher Scientific), with the Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific) or the TaqMan® Universal PCR Master Mix (Thermo Fisher Scientific).
-
18.
From this step on, the cells are producing viral particles and should be handled at biosafety level 2. All viral particles that are collected are also biosafety level 2.
-
19.
If your centrifuge is not able to run at 7000 × g, the centrifugation step can be carried out at lower g for a longer period of time (e.g., at 5000 × g for 30 min).
-
20.
We usually use one aliquot of viral particles to transduce one well of a 6- or 12-well plate. In order to optimize the viral transduction, it is recommended to determine the viral titer. Therefore, transduce cells with different volumes of the virus-containing supernatant and perform a qPCR on genomic DNA counting the number of integrated copies per cell.
-
21.
Directly after transfection, the iPSCs are biosafety level 2 and should be handled as such, after the medium change the next day, they are back at biosafety level 1.
-
22.
The calculation of the molecule number per μl in a solution with a concentration of 1 mg/ml is as follows: molecules/μl = (6.022 × 1023 1/mol [Avogadro constant]/(length of the plasmid × 660 Da [average molecular weight of a base pair])) × 10−6 g/μl. For the pAlbumin plasmid with a length of 7539 bp, the calculation is: (6.022 × 1023 1/mol/(7539 bp × 660 Da)) × 10−6 g/μl = 1.2 × 1011 molecules/μl.
-
23.
Avoid placing the PAM sequence into your sgRNA-expressing vector and the potential donor construct. It will be cut once sgRNA and Cas9 are expressed. The vectors from the Zhang lab can be ordered in different versions, that is, with GFP or puromycin expression. If positive cells should be sorted with flow cytometry, GFP is optimal. When expanding of single cells and subsequent picking of monoclonal colonies is preferred, use puromycin with the version V2 on Addgene, which is corrected from a previous version. The success rate of this cloning strategy is usually very high.
-
24.
One or two guanines can be added for more efficiency of the U6 promoter if the designed sgRNA is not starting with it. They have to be added to the bottom oligo as reverse complement in addition to the sgRNA sequence as well.
-
25.
The T7 endonuclease I assay is performed as follows: Transfect the sgRNA- and Cas9-expressing constructs into a test cell line (e.g., 293T/17, see Subheadings 2.3 and 3.3). Isolate the DNA using a DNA extraction kit, such as the DNeasy® Blood and Tissue Kit (Qiagen). Amplify the locus using flanking primers tested for specificity in advance. Purify the reaction using a PCR Purification kit (Qiagen). Elute in 30 μl. Mix 200 ng purified PCR product, 2 μl NEBuffer™ 2 and water to a total volume of 19 μl. Hybridize the PCR product in a thermocycler by heating to 95 °C and ramp down to 85 °C with −2 °C/s, then to 25 °C with 0.1 °C/s and hold at 4 °C. Add 1 μl T7 endonuclease I to the reaction and incubate at 37 °C for 20 min. Run on a 2% agarose gel (30 min, 90 V) to see if one or more bands appear. If two or three bands are visible, the sgRNA works fine. The T7 endonuclease cuts at wobbles that appear with reannealing of nonfitting DNA [13] strands. This happens if the Cas9 cuts parts of the DNA of the population of cells used as the test cell line.
-
26.
Rat Primary Cortical Astrocytes stick to the plastic used in cell culture dishes and centrifuge tubes. Prior to use, rinse all material that will come in contact with the cells with medium to prevent cells from sticking to the plastic.
-
27.
Since the cleaning of the coverslips is very time-consuming, it can also be done in 1 day. Briefly, rinse the coverslips two times in ddH2O, then add 50 ml 1 M HCl and shake for 1 h. Rinse three times with ddH2O by shaking for 2 min, and then rinse once more with ddH2O by shaking for 1 h. Shake three times in 100% ethanol for 2 min and one time for 1 h. Sterilize the coverslips at 225 °C for 2–3 h. Successful cleaning will be accompanied by an even spread of coating solution across the whole surface of the coverslip. If problems with adhesion occur, go back to the long protocol.
-
28.
The purpose of the spacers is to allow growth of the induced neurons in close proximity to the astrocyte feeder layer but without physical contact.
-
29.
The coverslips can have any size depending on the requirements of the experiment. We routinely use 12 mm coverslips equipped with three paraffin feet in a 24-well plate. It is recommended to add additional volume of medium to the well to completely cover the coverslips in order to avoid floating.
-
30.
We add approximately 50 μl ddH2O per week for a 24-well plate to compensate for volume loss due to evaporation. Store a test plate full of H2O in the incubator and weigh in weekly intervals to check for evaporation.
References
Srikanth P, Young-Pearse TL (2014) Stem cells on the brain: modeling neurodevelopmental and neurodegenerative diseases using human induced pluripotent stem cells. J Neurogenet 28:5–29
Shi Y, Inoue H, Wu JC, Yamanaka S (2017) Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 16:115–130
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096
Busskamp V et al (2014) Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol 10:760
Chanda S et al (2014) Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Reports 3:282–296
Yang N et al (2017) Generation of pure GABAergic neurons by transcription factor programming. Nat Methods 14:621–628
Zhang Y et al (2013) Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78:785–798
Barde I, Salmon P, Trono D (2010) Production and titration of lentiviral vectors. Curr Protoc Neurosci. Chapter 4, Unit 4 21
Zhao S et al (2016) PiggyBac transposon vectors: the tools of the human gene encoding. Transl Lung Cancer Res 5:120–125
Ran FA et al (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Hsu PD et al (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832
Guschin DY et al (2010) A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649:247–256
Sakuma T, Nakade S, Sakane Y, Suzuki KT, Yamamoto T (2016) MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc 11:118–133
Lam RS, Topfer FM, Wood PG, Busskamp V, Bamberg E (2017) Functional maturation of human stem cell-derived neurons in long-term cultures. PLoS One 12:e0169506
Kaech S, Banker G (2006) Culturing hippocampal neurons. Nat Protoc 1:2406–2415
Gibson DG (2011) Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361
Acknowledgments
This work was supported by Volkswagen Foundation Freigeist fellowship A110720 and ERC starting grant 678071—ProNeurons to V.B. E.J.S. and L.K.K. were supported by the DIGS-BB PhD Program.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, a link is provided to the Creative Commons license and any changes made are indicated.
The images or other third party material in this chapter are included in the work's Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work's Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2019 The Author(s)
About this protocol
Cite this protocol
Sauter, E.J., Kutsche, L.K., Klapper, S.D., Busskamp, V. (2019). Induced Neurons for the Study of Neurodegenerative and Neurodevelopmental Disorders. In: Ben-Yosef, D., Mayshar, Y. (eds) Fragile-X Syndrome. Methods in Molecular Biology, vol 1942. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-9080-1_9
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9080-1_9
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-9079-5
Online ISBN: 978-1-4939-9080-1
eBook Packages: Springer Protocols