Abstract
Background
Adiponectin plays key roles in regulating appetite and food intake. Altered circulating adiponectin levels have been observed in human eating disorders such as anorexia nervosa, bulimia nervosa or binge eating. In addition, an association between circulating adiponectin levels and human eating behaviour (EB) has been reported. Interestingly, a disturbance in eating behaviour is the defining characteristic of human eating disorders. However, it is unknown whether adiponectin is causally implicated in human EB. We therefore aimed to investigate the causal effect of adiponectin on EB.
Results
Mendelian randomization (MR) analysis estimated the influence of blood adiponectin on EB by combining data on the association of adiponectin gene (ADIPOQ) variants with adiponectin levels and with three EB factors involving disinhibition, restraint and hunger. Using inverse-variance weighted (IVW) regression method and other complementary MR techniques (weighted median regression, MR Egger and weighted modal regression), the MR analysis revealed a broadly consistent evidence that higher blood adiponectin concentration was significantly associated with increased EB factor disinhibition (beta coefficient for IVW regression [βIVW], 3.05; 95% confidence interval [CI] 1.10, 5.00) but non-significantly associated with increased EB factor restraint (βIVW, 0.17; 95% CI − 1.85, 2.18), and increased EB factor hunger (βIVW, 1.63; 95% CI − 0.75, 4.01).
Conclusions
Overall, our findings indicate a causal role of adiponectin levels in eating disinhibition but not in eating restraint and hunger.
Similar content being viewed by others
Background
Human circulating adiponectin is a well-described 30 kDa adipocytokine implicated in a wide range of anti-inflammatory [1], insulin-sensitizing [2] and eating disorder [3] pathways. Serum adiponectin levels are highly heritable [4,5,6,7] and, in contrast to adipokines, are inversely correlated with several cardiovascular risk factors such as obesity, type 2 diabetes mellitus, coronary artery disease and stroke [8,9,10,11,12,13]. Adiponectin serum levels may be influenced by nutritional compounds and probably increased food intake which may in turn serve as a positive feedback process [14]. Besides several other loci such as ARL15 (ADP-ribosylation factor-like 15 gene locus) [15], CDH13 (cadherin 13 gene locus) and KNG (kininogen gene locus) [16, 17], candidate and genome-wide association studies (GWAS) have shown pronounced associations between common polymorphisms in the adiponectin gene (ADIPOQ) and adiponectin levels [18,19,20,21]. Interestingly, polymorphisms of the ligand adiponectin gene, ADIPOQ, have been linked with a range of important clinical parameters such as body mass index (BMI), insulin resistance, cardiovascular disease and type 2 diabetes [22,23,24].
A number of observational studies have linked altered adiponectin levels with several eating disorders involving anorexia nervosa, bulimia nervosa and binge eating disorders [3]. In particular, several studies showed that serum adiponectin levels are increased in patients affected with anorexia nervosa [25,26,27] perhaps due in part to the lack of negative feedback exerted by fat mass in adiponectin production and/or enhanced insulin sensitivity [28]. In addition, studies have shown that serum adiponectin secretion in patients with bulimia nervosa could be the same, upregulated or downregulated when compared to those found in healthy controls. Moreover, the studies also indicated that adiponectin levels of bulimia nervosa patients were lower than those found in anorexia nervosa patients (reviewed in, e.g. [3]). Further, binge eating disorder was shown to be accompanied with lower serum adiponectin levels than in normal individuals [29, 30]. Of note, a disturbance in eating behaviour is the hallmark of the above clinical eating disorders. Patients with bulimia nervosa experience recurrent episodes of binge eating [31]. Patients with anorexia nervosa severely resist dietary intake and showed a persistent disturbance in eating behaviour even after restoration of body weight and significant improvements in eating disordered and psychological symptoms [32].
Mendelian randomization is an efficient analytical tool that uses genetic variants as instrumental variables to estimate the causal relationship between an exposure and outcome [33]. This method relies on the random assignment of genetic variants during gametogenesis to reduce the possibility of confounding [34, 35]. In addition, since genetic variant is established at conception (i.e. genotype being a fixed exposure), MR reduces the possibility of reverse causality [33]. Taken together, this confirms MR as an efficient tool that can substantially improve causal inference from observational data [36]. If adiponectin serum levels are altered in eating disorders, the genetic variant associated with adiponectin concentration should be associated with eating behaviour. We investigated this assumption through the use of a number of complementary MR techniques to examine the causal nature of the association between blood adiponectin level and three-factor eating behaviour involving restraint, disinhibition and hunger in Central European (CEU) population.
Methods
Data sources
Summary data on the association between ADIPOQ single-nucleotide polymorphisms (SNPs) and the phenotypes of interest were extracted from Rohde et al.’s paper (898 individuals of European ancestry) [37] and public databases of different consortia: ADIPOGen for adiponectin (29,347 individuals of European ancestry) [38] and GIANT (Genetic Investigation of ANthropometric Traits) for BMI (229,735 individuals) [39] and WC (795,447 individuals) [40].
Instrumental variables
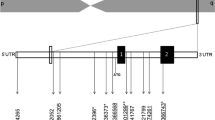
All the six but one ADIPOQ SNPs (Table 1) for our instrumental variable analyses were selected from 145 SNPs strongly (P < 5 × 10−8) associated with blood adiponectin levels in the European ancestry GWAS meta-analysis from the ADIPOGen consortium [38]. The remaining one SNP also selected from the ADIPOGen consortium was less strongly (P = 2.3 × 10−7) associated with adiponectin levels in the European ancestry GWAS meta-analysis [38]. We assessed correlations (linkage disequilibrium) among these SNPs using the LDlink [41] and found these SNPs to be independent variants.
Data analysis procedure
We performed a two-sample Mendelian randomization analysis using summary data from genome-wide association studies (GWAS). SNPs, previously reported to be associated with blood adiponectin levels, were used as instrumental variables for testing the causal effect of adiponectin on the three eating behaviour factors. Data on the association of ADIPOQ SNPs with (1) adiponectin levels (first sample) and (2) eating behaviour factors (second samples) were combined to estimate the influence of blood adiponectin on eating behaviour. First, we selected and obtained the beta coefficients and standard errors of six (6) GWAS significant SNPs that predicted adiponectin levels in the ADIPOGen consortium and were available in the eating behaviour outcome GWAS. Second, we re-analysed the summary data of these six SNPs on eating behaviour using their genotype frequencies, means and standard deviations as available in the Rohde et al.’s paper [37] using allelic model of inheritance to obtain the beta coefficients (β) and standard errors (SE) associated with the SNPs (Additional file 1: Table S1). These six ADIPOQ SNPs were then used for MR analysis. We also examined the standardized mean difference (and P values) of two eating behaviour risk factors involving BMI-adjusted WC and BMI per allele of the selected ADIPOQ SNPs for evidence of an effect of the SNPs on these risk factors. This was to enable us to assess the presence of potential bias (horizontal pleiotropy) or mediation of the effect of ADIPOQ SNPs with these eating behaviour risk factors.
Estimation of causal effect
The SNP-exposure and SNP-outcome associations for each of the eating behaviour factors were combined in a random effect meta-analysis using the inverse-variance weighted (IVW) regression method as described by Burgess et al. [42]. The β-coefficient of each eating behaviour factor per one natural log greater adiponectin level and its standard error (SE) were calculated.
Sensitivity analysis
While we considered our MR approach unlikely to be biased by horizontal pleiotropy given the functional relationship of ADIPOQ to adiponectin levels, yet, we investigated the presence of potential bias (horizontal pleiotropy), by running a number of sensitivity analyses involving weighted-median regression, MR Egger and weighted modal regression that provide causal estimates under less stringent assumptions than the traditional MR approach. The weighted median regression analyses can provide a consistent estimate for the true causal effect when up to half of the weights in the MR analysis are from ADIPOQ variants that exert pleiotropic effects on the eating behaviour factors [43, 44]. MR-Egger regression relaxes the assumption that the effects of our ADIPOQ variants on the eating behaviour factors are entirely mediated via the adiponectin concentration. This method allows for each ADIPOQ variant to exhibit some pleiotropy but assumes that each gene’s association with the adiponectin concentration is independent in magnitude from its pleiotropic effects (the InSIDE assumption) [45]. MR Egger achieved this by allowing an intercept term in the weighted regression analysis. The value of the intercept provides an estimate of the degree of pleiotropy affecting the result, while the beta (slope) coefficient represents the causal effect between adiponectin concentration and eating behaviour adjusted for pleiotropy. Finally, the weighted modal regression analyses relax our instrumental variable assumptions [46]. Analysis of association ADIPOQ SNPs with eating behaviour factors using an allelic model of inheritance was done in Statistical Analysis System (SAS) version 9.1.0 (SAS Stat) while MR analyses were implemented using MendelianRandomization package in R statistical software (R version 3.5.0).
Results
Association of genetic instruments with adiponectin, EB and EB risk factors
Figure 1 shows the associations of ADIPOQ SNPs, used as instrumental variables with adiponectin levels, EB and EB risk factors. Each adiponectin-increasing allele was associated with decreased EB factor restrain (beta coefficient [β], − 0.04; 95% confidence interval [CI] − 0.15, 0.08), increased EB factor disinhibition (β, 0.21; 95% CI 0.10, 0.33), and increased EB factor hunger (β, 0.13; 95% CI 0.02, 0.25). Of the six SNPs, there was some evidence of heterogeneity (P < 0.05) between studies that contributed to ADIPOGen consortium.
Forest plots of mean differences in log adiponectin levels and eating behaviour factors per allele of single nucleotide polymorphism (SNP). Analyses including six ADIPOQ SNPs associated with (1) adiponectin at genome-wide significant levels (P < 5 × 10−8) and (2) eating behaviour (EB) factors. Results for log adiponectin included 29,347 individuals from ADIPOGen Consortium and for EB factors included 898 individuals from Rohde et al. 2015*
None of the six ADIPOQ SNPs was associated with EB risk factors involving WC and BMI (Table 2). In addition, pooled effect estimates of these SNPs on these risk factors indicated no significant association with EB factors (Fig. 2). Finally, the adiponectin-increasing ADIPOQ variants were not associated with these available EB risk factors (Additional file 2: Table S2)
Forest plots of mean difference in eating behaviour risk factors per allele of single nucleotide polymorphism (SNP). Analysis including six ADIPOQ SNPs associated with adiponectin at genome-wide significant levels (P < 5 × 10−8). Results for BMI and WC included 229,735 and 795,447 individuals, respectively, from GIANT consortium
Effect of blood adiponectin concentration on EB
Table 3 shows the results of all MR analyses assessing the association of genetically predicted adiponectin with the three EB factors. Using IVW regression, higher adiponectin concentration was strongly and significantly associated with increased EB factor disinhibition (β, 3.05; 95% CI 1.10, 5.00). Similarly, using both weighted median MR and weighted modal MR, there was strong evidence of a significant association (weighted median β, 2.66; 95% CI 0.48, 4.84; weighted modal β, 2.63; 95% CI 0.54, 4.73). MR-Egger point estimate (− 0.32) was non-significant and in the opposite direction. The intercept indicated no evidence of pleiotropy (β, 0.23; 95% CI − 0.04, 0.51).
For EB factor restraint, higher adiponectin concentration was weakly and non-significantly associated with an increased likelihood of the EB (IVW β, 0.17; 95% CI − 1.85, 2.18). Results were similar when using weighted median MR (β, 0.98; 95% CI − 1.28, 3.24), MR Egger (β, − 3.85; 95% CI − 0.52, 8.21) and weighted modal estimate (β, 1.07; 95% CI − 1.12, 3.26). The intercept indicated no substantial evidence of pleiotropy (β, − 0.25; 95% CI − 0.53, 0.02).
Finally, there was evidence of an association between higher adiponectin concentration and increased hunger across all but one MR techniques. Inverse-variance weighted MR, weighted median MR and weighted modal MR all produced beta coefficients in the same direction. MR Egger on the other hand had a beta coefficient in the opposite direction. However, we found no statistical evidence of association for all methods (e.g. inverse-variance weighted β, 1.63; 95% CI − 0.75, 4.01). The intercept in the MR-Egger analysis did not indicate any substantial evidence of pleiotropy (β, 0.22; 95% CI − 0.12, 0.55).
Discussion
Using data from ADIPOGen consortium and eating behaviour GWAS study with information for up to 29,347 and 898 participants, respectively, we find evidence for a causal association between adiponectin levels and eating disinhibition but not for eating restraint and hunger. These findings suggest that the observational association between effect allele carriers in the ADIPOQ SNPs showing elevated adiponectin serum levels along with the tendency to frequently overeat (eating disinhibition), also confirmed in our datasets, may be causal. This effect was not due to horizontal pleiotropy based on our further investigation of the effect using alternative MR approaches (such as MR Egger, median, and mode estimators) and our assessment of the genetic variants with known and available potential confounders (such as WC and BMI).
The observational study of the role adiponectin in eating behaviour [37] indicated that while some ADIPOQ SNPs were related to disinhibition and hunger, none of the associations withstood Bonferroni corrections for multiple testing perhaps due to the relatively small sample size of the study. Taken advantage of the large GWAS consortium data from ADIPOGen, we have undertaken the first MR study of the causal effect of adiponectin on eating behaviour. We applied a rigorous analysis plan to assess the validity and consistency of our findings. This included (1) adopting a conservative approach in selecting SNPs for our instrumental variables, (2) the use of multiple eating behaviour phenotypes, (3) exploring different MR approaches to test the robustness of our results, (4) extensively investigating the presence of bias that may be due to horizontal pleiotropy, and (5) using a two-sample MR to avoid statistical overfitting in comparison to a one-sample MR setting and providing an opportunity to substantially increase our study statistical power.
Our findings reinforce and extend the earlier observation that the minor allele carrier of several ADIPOQ loci showed elevated adiponectin levels along with increased disinhibition [37]. Interestingly, eating disinhibition, correlated with high amount of food intake, was demonstrated to be strongly related to overeating without hunger feelings in certain situational circumstances [47, 48]. It is thus plausible that the increased food intake serves as a positive feedback mechanism. As adiponectin levels are known nutritional compound influencer [14], they can activate adenosine monophosphate-activated protein kinase (AMPK)-mediated signaling through adiponectin receptor binding in the hypothalamus region of animal models [49, 50]. This mechanism seems to alter energy expenditure to cause overeating and ultimately increased body weight [49, 50]. Of note, eating disinhibition has been most consistently reported to be related with increased BMI and obesity [51].
Some limitations of this study should be considered. First, not all the genome-wide significant SNPs that predicted adiponectin levels were available in the outcome GWAS we used. Thus, we were not necessarily capturing the full variance with the included variants. In addition, we were not able to test for effect modification by age and gender because of the use of summary data only. In observational studies, the role of age and gender as modifiable factors of eating behaviour is well recognized [52,53,54]. Significant differences between male and female adiponectin serum levels have also been reported [37, 55]. Moreover, and surprisingly too, we did not find a positive association between circulating adiponectin and disinhibition in the MR-Egger analysis. This is generally inconsistent with results from other MR techniques and may likely indicate a false-negative finding. Notably, MR-Egger regression analysis yields less precise estimates than other MR methods, owing to a power penalty [36]. Further, we were not able to specifically assess the causal effect of the biologically active and high molecular weight adiponectin in this study. Although we explored the violation of the assumptions of MR, we cannot completely rule out bias due to independence and exclusion restriction assumptions. However, when we tested the association of our genetic instrument with the two available potential EB risk factors (i.e. BMI and WC), the genetic instrument showed no significant association with both. These results strengthen our estimates of the effect of blood adiponectin concentration on EB. Finally, MR studies require large sample sizes partly due to the very small amount of variation in the exposure explained by genetic instruments [33]. Thus, while we believe our study was sufficiently powered to detect relatively small effects, our analysis may have been underpowered to detect, perhaps, very small causal effects operating at the extremes of adiponectin distribution in eating restraint or hunger.
Conclusion
In conclusion, our MR study reported a potential association between circulating adiponectin and eating disinhibition and could have potential implications on pathological disorders which include anorexia nervosa, bulimia nervosa and binge eating.
Availability of data and materials
Data on adiponectin have been contributed by ADIPOGen consortium and have been downloaded from https://www.mcgill.ca/genepi/adipogen-consortium. Data on eating behaviour factors have been contributed by Rohde and colleagues and have been extracted from Genes Nutr (2015) 10:1 (see reference [37]). Data on anthropometric traits have been contributed by Genetic Investigation of ANthropometric Traits (GIANT) consortium and have been downloaded from http://www.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files. All the data used are publicly available.
Abbreviations
- ADIPOQ:
-
Adiponectin gene
- AMPK:
-
Adenosine monophosphate-activated protein kinase
- ARL15:
-
ADP-ribosylation factor-like 15 gene locus
- BMI:
-
Body mass index
- CDH13:
-
Cadherin 13 gene locus
- CEU:
-
Central European
- CI:
-
Confidence interval
- EB:
-
Eating behaviour
- GIANT:
-
Genetic Investigation of Anthropometric Traits
- GWAS:
-
Genome-wide association studies
- IVW:
-
Inverse-variance weighted
- KNG:
-
Kininogen gene locus
- MR:
-
Mendelian randomization
- SAS:
-
Statistical analysis system
- SE:
-
Standard error
- SNP:
-
Single-nucleotide polymorphisms
- WC:
-
Waist circumference
References
Yamauchi T, Nio Y, Maki T, Kobayashi M, Takazawa T, Iwabu M et al (2007) Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions. Nat Med 13(3):332–339
Berg AH, Combs TP, Du X, Brownlee M, Scherer PE (2001) The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat Med 8:947–953
Khalil RB, El Hachem C (2014) Adiponectin in eating disorders. Eat Weight Disord 19(1):3–10
Comuzzie AG, Funahashi T, Sonnenberg G, Martin LJ, Jacob HJ et al (2001) The genetic basis of plasma variation in adiponectin, a global endophenotype for obesity and the metabolic syndrome. J Clin Endocrinol Metabol 86:4321–4325
Vasseur F, Helbecque N, Dina C, Lobbens S, Delannoy V et al (2002) Singlenucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet 11:2607–2614
Cesari M, Narkiewicz K, De Toni R, Aldighieri E, Williams CJ et al (2007) Heritability of plasma adiponectin levels and body mass index in twins. J Clin Endocrinol Metab 92:3082–3088
Liu PH, Jiang YD, Chen WJ, Chang CC, Lee TC et al (2008) Genetic and environmental influences on adiponectin, leptin, and BMI among adolescents in Taiwan: a multivariate twin/sibling analysis. Twin Res Hum Genet 11:495–504
Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J et al (1999) Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun 1:79–83
Cnop M, Havel PJ, Utzschneider KM, Carr DB, Sinha MK, Boyko EJ et al (2003) Relationship of adiponectin to body fat distribution, insulin sensitivity and plasma lipoproteins: evidence for independent roles of age and sex. Diabetologia 46:459–469
Hivert MF, Sullivan LM, Fox CS, Nathan DM, D’Agostino RB Sr et al (2008) Associations of adiponectin, resistin, and tumor necrosis factor-alpha with insulin resistance. J Clin Endocrinol Metab 93:3165–3172
Tilg H, Moschen AR (2006) Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat Rev Immunol 6:772–783
Pischon T, Girman CJ, Hotamisligil GS, Rifai N, Hu FB et al (2004) Plasma adiponectin levels and risk of myocardial infarction in men. JAMA 291:1730–1737
Li S, Shin HJ, Ding EL, van Dam RM (2009) Adiponectin levels and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA 302:179–188
Mantzoros CS, Williams CJ, Manson JE, Meigs JB, Hu FB (2006) Adherence to the Mediterranean dietary pattern is positively associated with plasma adiponectin concentrations in diabetic women. Am J Clin Nutr 84:328–335
Richards JB, Waterworth D, O’Rahilly S, Hivert MF, Loos RJ et al (2009) A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet 5:e1000768
Jee SH, Sull JW, Lee JE, Shin C, Park J et al (2010) Adiponectin concentrations: a genome-wide association study. Am J Hum Genet 87:545–552
Wu Y, Li Y, Lange EM, Croteau-Chonka DC, Kuzawa CW et al (2010) Genome-wide association study for adiponectin levels in Filipino women identifies CDH13 and a novel uncommon haplotype at KNG1-ADIPOQ. Hum Mol Genet 19:4955–4964
Menzaghi C, Trischitta V, Doria A (2007) Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes 56:1198–1209
Hivert MF, Manning AK, McAteer JB, Florez JC, Dupuis J et al (2008) Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes 57:3353–3359
Ling H, Waterworth DM, Stirnadel HA, Pollin TI, Barter PJ et al (2009) Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS study. Obesity (Silver Spring) 17:737–744
Heid IM, Henneman P, Hicks A, Coassin S, Winkler T et al (2010) Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis 208:412–420
Ogundele OE, Adekoya KO, Osinubi AA, Awofala AA, Oboh BO (2018) Association of adiponectin gene (ADIPOQ) polymorphisms with measures of obesity in Nigerian young adults. Egypt J Med Hum Genet 19:123–127
Stumvoll M, Tschritter O, Fritsche A et al (2002) Association of the T-G polymorphism in adiponectin (exon 2) with obesity and insulin sensitivity: interaction with family history of type 2 diabetes. Diabetes 51(1):37–41
Vasseur F, Helbecque N, Lobbens S et al (2005) Hypoadiponectinaemia and high risk of type2 diabetes are associated with adiponectin-encoding (ACDC) gene promoter variants in morbid obesity: evidence for a role of ACDC in diabesity. Diabetologia 48(5):892–899
Modan-Moses D, Stein D, Pariente C, Yaroslavsky A, Ram A, Faigin M et al (2007) Modulation of adiponectin and leptin during refeeding of female anorexia nervosa patients. J Clin Endocrinol Metab 5:1843–1847
Pannacciulli N, Vettor R, Milan G, Granzotto M, Catucci A, Federspil G et al (2003) Anorexia nervosa is characterized by increased adiponectin plasma levels and reduced nonoxidative glucose metabolism. J Clin Endocrinol Metab 4:1748–1752
Terra X, Auguet T, Agüera Z, Quesada IM, Orellana-Gavaldà JM, Aguilar C et al (2013) Adipocytokine levels in women with anorexia nervosa. Relationship with weight restoration and disease duration. Int J Eat Disord 8:855–861
Delporte ML, Brichard SM, Hermans MP, Beguin C, Lambert M (2003) Hyperadiponectinaemia in anorexia nervosa. Clin Endocrinol (Oxf) 58:22–29
Monteleone P, Fabrazzo M, Martiadis V, Fuschino A, Serritella C, Milici N et al (2003) Opposite changes in circulating adiponectin in women with bulimia nervosa or binge eating disorder. J Clin Endocrinol Metab 11:5387–5391
Carnier J, Sanches Pde L, da Silva PL, de Piano A, Tock L, Campos RM et al (2012) Obese adolescents with eating disorders: analysis of metabolic and inflammatory states. Physiol Behav 105(2):175–180
Walsh DT (2011) The importance of eating behaviour in eating disorders. Physiol Behav 104(4):525–529
Sysko R, Walsh BT, Schebendach J, Wilson GT (2005) Eating behaviour among women with anorexia nervosa. Am J Clin Nutr 82(2):296–301
Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G (2008) Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 27:1133–1163
Burgess S, Butterworth A, Malarstig A, Thompson SG (2012) Use of Mendelian randomisation to assess potential benefit of clinical intervention. BMJ 345:e7325–e7325
Smith GD, Ebrahim S (2003) Mendelian randomization: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32:1–22
Davies NM, Holmes MV, Smith GD (2018) Reading Mendelian randomization studies: a guide, glossary and checklist for clinicians. BMJ 362:k601
Rohde K, Keller M, Horstmann A, Liu X, Eichelmann F, Stumvoll M et al (2015) Role of genetic variants in ADIPOQ in human eating behavior. Genes Nutr 10:1–10
Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA et al (2012) DIAGRAM+ Consortium; MAGIC Consortium; GLGC Investigators; MuTHER Consortium; DIAGRAM Consortium; GIANT Consortium; Global B Pgen Consortium; Procardis Consortium; MAGIC investigators; GLGC Consortium. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet 8:e1002607
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR et al (2015) LifeLines Cohort Study; ADIPOGen Consortium; AGEN-BMI Working Group; CARDIOGRAMplusC4D Consortium; CKDGen Consortium; GLGC; ICBP; MAGIC Investigators; MuTHER Consortium; MIGen Consortium; PAGE Consortium; ReproGen Consortium; GENIE Consortium; International Endogene Consortium. Genetic studies of body mass index yield new insights for obesity biology. Nature 518:197–206
Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Mägi R et al (2015) ADIPOGen Consortium, CARDIOGRAMplusC4D Consortium, CKDGen Consortium, GEFOS Consortium, GENIE Consortium, GLGC, ICBP, International Endogene Consortium, LifeLines Cohort Study, MAGIC Investigators, MuTHER Consortium, PAGE Consortium, ReproGen Consortium. New genetic loci link adipose and insulin biology to body fat distribution. Nature 518:187–196
Machiela MJ, Chanock SJ (2015) LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31(21):3555–3557
Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37:658–665
Bowden J, Davey Smith G, Haycock PC, Burgess S (2016) consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40:304–314
Kang H, Zhang A, Cai TT, Small D (2016) Instrumental variables estimation with some invalid instruments and its application to Mendelian randomization. J Am Stat Assoc 111:132–144
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44:512–525
Hartwig FP, Davey Smith G, Bowden J (2017) Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 46:1985–1998
Dykes J, Brunner EJ, Martikainen PT, Wardle J (2004) Socioeconomic gradient in body size and obesity among women: the role of dietary restraint, disinhibition and hunger in the Whitehall II study. Int J Obes. 28(2):262–268
French S, Jeffery R, Wing RR (1994) Food intake and physical activity: a comparison of three measures of dieting. Addict Behav 4:401–409
Kubota N, Yano W, Kubota T, Yamauchi T, Itoh S, Kumagai H et al (2007) Adiponectin stimulates AMP-activated protein kinase in the hypothalamus and increases food intake. Cell Metab 1:55–68
Minokoshi Y, Shiuchi T, Lee S, Suzuki A, Okamoto S (2008) Role of hypothalamic AMP-kinase in food intake regulation. Nutrition 9:786–790
Bryant EJ, King NA, Blundell JE (2008) Disinhibition: its effects on appetite and weight regulation. Obes Rev 5:409–419
Hays NP, Roberts SB (2008) Aspects of eating behaviors “disinhibition” and “restraint” are related to weight gain and BMI in women. Obesity 1:52–58
Jastreboff AM, Gaiser EC, Gu P, Sinha R (2014) Sex differences in the association between dietary restraint, insulin resistance and obesity. Eat Behav 2:286–290
Dakanalis A, Zanetti MA, Clerici M, Madeddu F, Riva G, Caccialanza R (2013) Italian version of the Dutch Eating Behavior Questionnaire. Psychometric proprieties and measurement invariance across sex, BMI-status and age. Appetite 71:187–1895
Heid IM (2006) Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes 2:375–384
Acknowledgements
We thank those authors whose data were used in this study for making their genome-wide data available to scientists. Those people acknowledged here may not necessarily agree with comments made in this article, and the authors take full responsibility for the contents of this article.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AAA conceived the study and the analysis plan, participated in its design and coordination, performed the statistical analyses and drafted the manuscript. OEO participated in the design of the study, assisted in the data acquisition (from public databases) and helped to draft the manuscript. KOA participated in study supervision and coordination, analysis plan and data interpretation. SAO carried out data acquisition and assisted in the statistical analyses. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Estimated Standardized Mean Difference and Standard Errors of EB Factors Per Allele of SNPs Based on Allelic Model of Inheritance on Summary Data from Rohde et al paper [37].
Additional file 2: Table S2.
Standardized Mean Difference (and 95% Confidence Interval [CI]) in Eating Behaviour Risk Biomarkers Per 1 U Increase in Genetically Instrumented Log Adiponectin Levels.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Awofala, A.A., Ogundele, O.E., Adekoya, K.O. et al. Adiponectin and human eating behaviour: a Mendelian randomization study. Egypt J Med Hum Genet 20, 17 (2019). https://doi.org/10.1186/s43042-019-0022-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-019-0022-5