Abstract
Background
Information on above-ground biomass (AGB) is important for managing forest resource use at local levels, land management planning at regional levels, and carbon emissions reporting at national and international levels. In many tropical developing countries, this information may be unreliable or at a scale too coarse for use at local levels. There is a vital need to provide estimates of AGB with quantifiable uncertainty that can facilitate land use management and policy development improvements. Model-based methods provide an efficient framework to estimate AGB.
Methods
Using National Forest Inventory (NFI) data for a ~1,000,000 ha study area in the miombo ecoregion, Zambia, we estimated AGB using predicted canopy cover, environmental data, disturbance data, and Landsat 8 OLI satellite imagery. We assessed different combinations of these datasets using three models, a semiparametric generalized additive model (GAM) and two nonlinear models (sigmoidal and exponential), employing a genetic algorithm for variable selection that minimized root mean square prediction error (RMSPE), calculated through cross-validation. We compared model fit statistics to a null model as a baseline estimation method. Using bootstrap resampling methods, we calculated 95 % confidence intervals for each model and compared results to a simple estimate of mean AGB from the NFI ground plot data.
Results
Canopy cover, soil moisture, and vegetation indices were consistently selected as predictor variables. The sigmoidal model and the GAM performed similarly; for both models the RMSPE was ~36.8 tonnes per hectare (i.e., 57 % of the mean). However, the sigmoidal model was approximately 30 % more efficient than the GAM, assessed using bootstrapped variance estimates relative to a null model. After selecting the sigmoidal model, we estimated total AGB for the study area at 64,526,209 tonnes (+/− 477,730), with a confidence interval 20 times more precise than a simple design-based estimate.
Conclusions
Our findings demonstrate that NFI data may be combined with freely available satellite imagery and soils data to estimate total AGB with quantifiable uncertainty, while also providing spatially explicit AGB maps useful for management, planning, and reporting purposes.
Similar content being viewed by others
Background
Information on forest resources in tropical developing countries is challenging to acquire due to financial and logistical constraints (Holmgren and Marklund 2007; Skutsch and Ba 2010; Stringer et al. 2012). However, tropical forest resources are being exploited and converted to other uses at rates that seem to outpace the capacity for regrowth (Shearman et al. 2012; Brink et al. 2014; Sawe et al. 2014; Suberu et al. 2014). This is especially true in the miombo ecoregion of southern Africa (Cabral et al. 2010; Mayes et al. 2015), where forest productivity is marginal (Frost 1996). In this context, forest monitoring programs are required to provide information on forest resources for: i) planning use and management activities at local levels (Stringer et al. 2012); and ii) designing effective policies and measures at national levels. Above-ground biomass (AGB) is a key variable of interest in forest monitoring programs, where estimates of AGB are necessary for assessing fuel wood and timber availability, as well as for monitoring forest carbon stocks. Integrating ground plot data from a forest inventory with environmental and/or remotely-sensed predictor variables to predict AGB has shown increasing utility for estimating AGB (Moisen et al. 2006; McRoberts et al. 2010; Lu et al. 2016; GOFC-GOLD 2015).
Optical remotely-sensed data alone have been used to estimate miombo AGB with varying success (Samimi and Kraus 2004; Kashindye et al. 2013; Næsset et al. 2016). The use of these data is complicated by complex, nonlinear relationships between AGB and vegetation indices (Lu et al. 2016). Active sensor remotely-sensed data such as Airbone Laser Scanning (ALS) or radar may provide improved accuracy of AGB models (Ryan et al. 2012; Mitchard et al. 2013; Mauya et al. 2015; Næsset et al. 2016), although this is not guaranteed (Solberg et al. 2015). Another way to improve AGB estimation is to incorporate percent canopy cover (CC) as a predictor variable (Tiwari and Singh 1984; Lefsky et al. 2002; Hall et al. 2006; González-Roglich et al. 2014; GOFC-GOLD 2015), particularly since percent canopy cover is more readily estimated by optical remotely-sensed data (Lefsky and Cohen 2003; Lu et al. 2016).
González-Roglich and Swenson (2016) capitalized on the relationship between AGB and CC for modeling forest carbon in temperate savannahs of South America. They first developed a model to estimate CC using Landsat 5 Thematic Mapper along with topographic and climatic variables, and then used the estimated CC as the sole predictor to estimate forest carbon. Using this process, the percent root mean square error (RMSE) for prediction of forest carbon was 35 %. Similar studies in arid woodlands of Australia (Suganuma et al. 2006) and Sudan (Wu et al. 2013) also reported favorable results using estimated CC to estimate AGB. One complication is that the definition of CC varies (Jennings et al. 1999). In some cases, CC was defined as the percentage of the sky blocked by tree crowns over a hemispherical view (Jennings et al. 1999), often measured using a spherical densiometer. Alternatively, the vertical projection of tree crowns onto the ground is currently used in many countries as the basis for minimum forest area definition (FAO 2014). González-Roglich and Swenson (2016) used the former definition, while Suganuma et al. (2006) and Wu et al. (2013) used the latter. Therefore, it is an open question as to which measure of CC may better relate to AGB.
Models to estimate AGB that also incorporate information on topography, soils, and disturbances may further improve the accuracy (Moisen and Frescino 2002; Powell et al. 2010; Pflugmacher et al. 2014; GOFC-GOLD 2015). In the miombo ecoregion, as in other ecosystems, ecological and physiological factors limit vegetation establishment and survival, while disturbances result in changes to vegetation structure and composition (Frost 1996; Chidumayo et al. 1996; Sankaran et al. 2008). Soil characteristics, fire regimes, and anthropogenic use have all been identified as principal determinants of vegetation structure (Timberlake and Chidumayo 2011; Ryan and Williams 2011). Investigations of these and other variables for estimating miombo AGB appear to be sparse, and further exploration is warranted.
The main goal of this research is to investigate methods for estimating total AGB for a study area within the miombo ecoregion by using ecologically relevant predictor variables. For this purpose, we compared models established within a model-based framework, where inference is derived from the assumption that the y i observations of AGB are considered realizations of the random variable Y i , which in turn are realizations of a random process termed a superpopulation (Gregoire 1998; Ståhl et al. 2016). To assess these models, we compared outcomes to a simple design-based estimate commonly used in forest inventories. The design-based estimate assumes that the population of Y i is fixed, not random, while the sample of y i observations is the realization of a random process, and inference is independent from assumptions regarding the probability distribution of Y i in the population (Gregoire 1998; Ståhl et al. 2016). In this framework, we then addressed the following specific questions: 1) Which definition of CC is more useful as a single predictor variable to estimate AGB, the hemispherical view or the vertical projection view? 2) Does AGB estimation accuracy improve if CC is combined with other predictor variables? 3) What model-based AGB estimation method performs best in terms of fit and validation statistics? and 4) How do model-based methods compare to a simple design-based estimate of the sample mean? This research is intended as a case study to help inform development of forest monitoring programs which apply National Forest Inventory (NFI) data to estimation of AGB at sub-national or national scales.
Methods
Study area
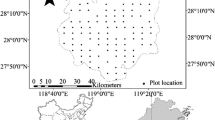
This study was carried out in Nyimba District (14°-15°S, 30°-31°E, ~1,000,000 ha), Eastern Province, Zambia (Fig. 1). Nyimba lies in the center of the miombo ecoregion, a biome of diverse vegetation types dominated by tree species from the Caesalpinioideade sub-family of leguminous plants (Timberlake and Chidumayo 2011). Across the ecoregion, vegetation composition and structure varies depending on climate, soil, landscape position, and level of disturbance (Frost 1996; Timberlake and Chidumayo 2011). Nyimba is found within the dry miombo ecozone and is dominated by four vegetation types (Table 1; GRZ 1976; Timberlake and Chidumayo 2011). Approximately 75 % of the district can be characterized as forest land, based on canopy cover ≥ 10 % per 0.5 ha area (Halperin et al. 2016). Nonforest land uses are dominated by small-scale agriculture (Gumbo et al. 2016). Fire is a frequent disturbance, with an estimated return interval of one to three years (Frost 1996; Timberlake and Chidumayo 2011). Fires generally result from people engaged in agricultural land clearing, charcoal making, and hunting, with early dry-season fires exhibiting less intensity than late dry-season fires (Frost 1996). Average rainfall is approximately 600–900 mm per year (GRZ 1968); most rain falls in the December to February period, with a distinct dry season from May to November. Soils are generally characterized as Lithosol-Cambisols on the hills and plateaus, and Fluvisol-Vertisols in the valleys (GRZ 1986). Elevation ranges from 450 m along the Luangwa River valley bottom to 1000 m on the plateau area around Nyimba district capital, and higher on the mountain tops in the western half of the district.
Ground plot measurements
Ground plot measurements were collected at 64, 0.1 ha permanent plots from May – December 2013 (Fig. 1). The ground plots were located on a systematic 10 km × 10 km grid corresponding to the Zambian Integrated Land Use Assessment 2 (ILUA2), an NFI program implemented by the Zambian Forest Department in collaboration with the FAO (GRZ 2014). Nationally, the Zambian Forest Department collects field data at approximately 20 % of the grid intersections using a cluster plot design. Each cluster plot has four 20 m × 50 m rectangular subplots, and each subplot has a nested 10 m × 20 m microplot starting at the same origin as the subplot. In this study, we collected data only for the first ground subplot of a cluster to increase the number of locations within time constraints, thereby capturing variations in conditions and land uses across the landscape. As such, we refer to the 20 m × 50 m area where we collected ground measurements as the plot, and the 10 m × 20 m area as the subplot.
Measurements within each ground plot followed the ILUA2 protocol (GRZ 2014). Species, total height, and diameter at breast height (DBH; outside bark diameter at 1.3 m above ground) were measured and recorded for each live tree with DBH ≥ 10 cm at the plot level and each live tree ≥ 5 cm and < 10 cm DBH at the subplot level. These tree attributes were used to calculate tree-level AGB, using species-specific allometric models developed by Chidumayo (2012) and utilized by the Zambian Forest Department (GRZ 2014). In some cases, species-specific models were not available and forest type models were used (Chidumayo 2012). We summarized tree-level AGB in plots and subplots to obtain plot-level AGB in tonnes per hectare (t∙ha−1).
We also measured percent CC for each ground plot using a spherical densiometer and recorded the dominant vegetation type (i.e., miombo woodland, mopane woodland, munga woodland, or riparian forest, GRZ 2014). We recorded ‘nonforest’ if the ground plot was dominated by nonforest land use with none of the vegetation types present. Ground plot coordinates were determined using a Trimble GeoXT, which were differentially corrected (+/− 1 m). Lastly, we gathered GPS line data for roads and point data for village locations. Most of the ground plots (49) were visited between 5 May and 30 July 2013, while the remaining plots (15) were visited between 27 Oct and 6 Dec 2013.
Predictor variables
We employed two types of estimated CC as a single predictor variable in separate models to estimate AGB. First, CC was estimated as the vertical projection of tree crowns onto the ground using a binomial generalized additive model (GAM) model developed for the same study area (Halperin et al. 2016), referred to as \( {\hat{CC}}_{\mathrm{VERT}} \). Second, CC was estimated as the percentage of the sky blocked by tree crowns over a hemispherical view by using the densiometer. For this purpose, we built a binomial GAM to estimate percent CC using the same predictor variable selection methods described in Halperin et al. (2016), and refer to its estimates as \( {\hat{CC}}_{\mathrm{HEMI}} \). We compared the results of estimating AGB using either \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \), and used the better performing model as the base upon which to add other predictor variables.
Disturbance factors, such as small scale conversion to agriculture, unmanaged harvesting for charcoal and timber, and fire are known to affect miombo AGB (Chidumayo 1988; Frost 1996; Timberlake and Chidumayo 2011). Proximity to roads and villages has been shown to be related to increasing levels of anthropogenic disturbance and decreased AGB (Helmer et al. 2008; Ahrends et al. 2010). We calculated Euclidean distance from each ground plot center to roads and to villages, based on the GPS line and point data collected during field surveys (Table 2). Fire frequency and intensity both impact miombo AGB, where low intensity burns are more characteristic of the early dry season when grass is still curing, and higher intensity burns are more likely to occur in the late dry season (Ryan and Williams 2011). We included monthly fire history data for 500 m pixels for 2003–2013, derived from the MODIS Burned Area Product (Boschetti et al. 2013). We processed the fire history data and determined the number of low intensity, early season fires and high intensity, late season fires per pixel (Table 2).
Underlying factors that affect miombo AGB include topography and soils (Frost 1996; Timberlake and Chidumayo 2011). We assessed landscape position using the compound topographic index (CTI), distance to perennial rivers, and slope percent. These variables were calculated based on 30 m Shuttle Radar Topography Mission Digital Elevation Model (USGS 2004) (Table 2). ISRIC – World Soil Information has produced a consistent and comprehensive data source for soils information at 250 m pixel size, covering the continent of Africa (Hengl et al. 2015). The data is provided as estimates at six depths (2.5, 10, 22.5, 45, 80, and 150 cm). We downloaded these data, clipped each dataset to the study area boundary, and calculated the mean across all depths for each variable (Table 2).
The use of predictor variables developed from optical satellite imagery in models to estimate AGB has a long history. However, the utility of various bands, indices, or texture values to use as predictor variables still remains an area of active research (Banskota et al. 2014; Lu et al. 2016). We downloaded Landsat 8 OLI data from the Landsat Ecosystem Disturbance Adaptive Process (LEDAPS) archive that spatially and temporally corresponded to the study area and the ground measurements (Path 170, Row 70; Path 171, Row 70). LEDAPS data are processed to surface reflectance (Masek et al. 2006); therefore, we did not perform atmospheric correction or radiometric normalization. We created one mosaic from the two scenes that were found to be in acceptable agreement with the GPS line data for roads based on visual assessment. We derived 24 predictor variables from Landsat 8 OLI satellite imagery in order to assess the use of a wide range of spectral and textural attributes for predicting AGB (Table 3).
Before selecting pixels to match with ground plots for all predictor datasets, we masked out major rivers and paved roads by on-screen digitizing using Landsat data at 1:10,000. All spatial data were clipped to the study area boundary. To account for spatial extent mismatches between ground plot size and predictor variables, we resampled all predictor variable spatial data to a 20 m × 50 m pixel size using a nearest neighbor method which maintained the actual pixel values. The values for each predictor variable were assigned to ground plots, based on proximity of the ground plot center to the nearest pixel center for each predictor variable. Preparation of spatial predictor variables was conducted using the R raster package v.2.3-41 (Hijmans et al. 2015) and ArcGIS v. 10.1 (ESRI 2012).
AGB Models
Models using CC only
As noted, we estimated AGB first using either \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \) as a single predictor variable using three models. A semiparametric GAM was used since it is well-suited to modeling complex relationships where an underlying nonlinear relationship is not known a priori (Wood 2006). Because of their flexibility, GAMs have been used to estimate forest attributes from remotely-sensed data (Moisen and Frescino 2002; Kattenborn et al. 2015). The form of the GAM is:
where i is the ground plot, AGB i is in t∙ha−1, β0 is the intercept, ε i is the residual error, and f j is the smoothing function for a predictor variable x j created via a regression spline (Wood 2006). The GAM was fit with a normal distribution, the log link, and the default variance function. Using the log link avoids negative AGB estimates. A value of 0.1 was added to the AGB for two ground plots with no trees. We found that setting the maximum smoothing parameter at 2 was sufficiently flexible to account for potential nonlinearity in the model, yet reduce the possibility of overfitting that may occur in GAM’s (Moisen et al. 2006; Halperin et al. 2016). The GAM was fit with the mgcv package v. 1.8-9 in R v. 3.2.2 (Wood 2015; R Development Core Team 2015).
We also fit two nonlinear models that restricted the AGB estimate to be positive with an upper limit. First, we used a sigmoidal model (e.g., McRoberts et al. 2015):
where parameters are α and the β’s, and the other terms are as in Eq. 1. Second, we used an exponential model, also used for estimating AGB (e.g., Anaya et al. 2009):
To fit these nonlinear models, we used the Gauss-Newton method in the nls function in R v.3.2.2. Starting parameters were estimated using the linearized form of each nonlinear model (i.e., log (AGB i )). After experimenting with a flexible or fixed asymptote α, we found that setting α = 500 produced realistic estimates. This is in line with Næsset et al. (2016), who truncated miombo AGB estimates from linear ordinary least squares models at twice the maximum observed value in their sample data. For each fitted model (three models and \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \)), we used 20-fold cross-validation and calculated the root mean square prediction error (RMSPE) and RMSPE as a percent of the mean (RMSPE%), defined as:
where y iv is the AGB (t∙ha−1) of ground plot i not used for fitting in fold v, \( {\hat{y}}_{iv} \) is its corresponding estimate, and n = the number of ground plots (64). RMSPE% was calculated as:
where \( \overline{y} \) is the mean AGB (t∙ha−1) calculated from the ground plots. We then calculated and compared the mean RMSPE and mean RMSPE% across the three models using \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \). Based on these validation statistics, we included either \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \) in models with multiple predictor variables.
Models using multiple predictor variables
Next, we investigated whether expanding the models with increasingly diverse combinations of predictor variable sets could improve the accuracy of AGB estimates. For this purpose, we compared models using: \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \) alone; remotely-sensed (Landsat) variables alone; environmental and disturbance (Env/Dist) variables alone; and combinations of each predictor variable set (i.e., Landsat and Env/Dist, \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \) and Landsat, etc.). A genetic algorithm (GA) was used to perform predictor variable selection within each predictor variable set. The GA has been demonstrated to provide near optimality for predictor variable identification and subsetting when there are a large, diverse range of possible predictor variables (Tomppo and Halme 2004; Garcia-Gutierrez et al. 2014); a description of the algorithm is given in Scrucca (2013). We set the objective criteria (i.e., “fitness”) to minimize the RMSPE, and established the GA stopping criteria at 20 consecutive generations with no improvement in fitness, or a maximum of 100 generations. We restricted the number of variables under consideration to a maximum of six in each predictor variable set, in order to decrease the risk of near multicollinearity and overfitting. For each predictor variable set (except \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \) alone), we implemented the GA one time and recorded the lowest RMSPE. We then calculated and compared mean RMSPE and mean RMSPE% for each predictor variable set, and chose the predictor variable set that minimized these two fit statistics. The GA analysis was conducted using the GA package v.2.2 in R (Scrucca 2013).
Final variable selection
We performed final predictor variable selection using the predictor variable set that minimized RMSPE in the previous section. For this purpose, we implemented the GA 100 times for each model using the same fitness and stopping criteria as before, and selected the subset of predictor variables that minimized RMSPE across all 100 GA implementations. For the sigmoidal and exponential models, we also considered commonly used transformations (i.e., square, square root, inverse, etc.) for each predictor variable, once the predictor variables were chosen through this process. Use of transformations for predictor variables has been examined in nonlinear models for improving model performance (Wang et al. 2007; Timilsina and Staudhammer 2012). To compare performance among the three models, we computed the following fit statistics: root mean square error (RMSE), RMSE as a percent of the mean (RMSE%), mean difference (MD), and Pseudo-R2 as follows:
where y i and \( {\hat{y}}_i \) are the observed and estimated AGB (t∙ha−1) for ground plot i, and n and \( \overline{y} \) were defined in Eq. 4 and Eq. 5, respectively. We also calculated 20-fold validation statistics, namely RMSPE (Eq. 4) and the mean prediction difference (MPD) defined as:
where y iv , \( {\hat{y}}_{iv} \), and n were defined for Eq. 4.
Uncertainty assessment and model selection
After choosing the best performing predictor variable subset for each model, we estimated the AGB total (tonnes), 95 % confidence interval (CI) for the total, and AGB (t∙ha−1) for the entire study area. We accomplished this using a bootstrap method involving resampling n samples from the ground plot data with replacement using equal probability sampling, following recommendations from McRoberts et al. (2011) and based on work by Efron and Tibshirani (1986). Using 500 bootstrap samples, we refitted each model to the predictor variable data covering the entire study area, and estimated AGB for every 20 m × 50 m pixel. Because the GAM is not asymptotic, we limited the maximum estimated value for AGB (t∙ha−1) to the same value as the asymptote for the sigmoidal and exponential models, as is commonly done in non-asymptotic models (e.g., Næsset et al. 2016). For each model, we estimated total AGB (\( {\hat{\mathrm{Y}}}_{\mathrm{boot}} \)) using the following equation:
where \( {\hat{\mathrm{Y}}}_{\mathrm{boot}}^{\mathrm{k}} \) is the estimated total AGB by summing all pixels (in tonnes) across the study area for bootstrap k, and nboot is 500. AGB/ha was calculated from dividing \( {\hat{\mathrm{Y}}}_{\mathrm{boot}} \) by the number of ha in the study area. We estimated the variance for \( {\hat{\mathrm{Y}}}_{\mathrm{boot}} \) using the following equation:
We used this variance estimate to calculate a 95 % confidence interval (CI) for estimated total AGB based on the normal distribution (Efron and Tibshirani 1986), and reported the half-width of the CI for each model. To establish a baseline for comparing model performance, we fit a null model with no predictor variables. Using this null model, we estimated the AGB total, 95 % CI for the total, and AGB (t∙ha−1) (Bater et al. 2009). Then, we calculated the relative efficiency for each model by dividing the null model variance estimate by each model’s variance estimate, following Næsset et al. (2016). Finally, we compared the four model-based estimates of total AGB, AGB/ha, and CI to a simple design-based estimate of AGB total, 95 % CI for the total, and AGB (t∙ha−1) using the NFI systematic sample of ground plots, assuming equal probability sampling (Cochran 1977).
Based on these statistics, along with observed versus predicted AGB graphs, we chose one model and used this to estimate AGB for each 20 m × 50 m pixel in the study area. To assess the pixel-level variability of the AGB estimates, we calculated the Coefficient of Variation (CV) in percent (i.e., standard deviation per pixel across the 500 resamples divided by the corresponding mean pixel value, multiplied by 100, which was also used by Carreiras et al. 2013; Faßnacht et al. 2014; Kattenborn et al. 2015), present a map of CV, and report the mean CV for the study area.
Results
Models using CC only
Observations of AGB (t∙ha−1) and CCVERT (%) for the miombo woodland vegetation type follow a more regular pattern than the mopane woodland and riparian forest vegetation types, which exhibit a large range of observed AGB values at low observed values of CCVERT (Fig. 2). Observations of CCHEMI greater than 50 % display a wide range of observed AGB values; however, observed AGB values corresponding to CCHEMI less than 50 % follow a more regular pattern. As expected, the six nonforest ground plots which were measured have low observed AGB and low observed CC for both forms of CC. A binomial GAM was used to fit separate models for CCVERT and CCHEMI; the model to estimate \( {\hat{CC}}_{\mathrm{VERT}} \) had better fit and validation statistics than the model to estimate \( {\hat{CC}}_{\mathrm{HEMI}} \) (Table 4). However, in predicting AGB, clear differences are apparent in the usefulness of CC as the percentage of the sky blocked by tree crowns over a hemispherical view (\( {\hat{CC}}_{\mathrm{HEMI}} \)) versus CC as the vertical projection of tree crowns onto the ground (\( {\hat{CC}}_{\mathrm{VERT}} \)). Across three model forms, \( {\hat{CC}}_{\mathrm{HEMI}} \) had an average RMSPE% that was 18 % lower than for \( {\hat{CC}}_{\mathrm{VERT}} \) (Table 5). The exponential model performed poorly with RMSPE (72.1 t∙ha−1) larger than the mean estimate of AGB t∙ha−1 from the sample data (64.1 t∙ha−1). Considering only the GAM and sigmoidal models, the average difference in RMSPE between \( {\hat{CC}}_{\mathrm{VERT}} \) and \( {\hat{CC}}_{\mathrm{HEMI}} \) as a single predictor variable was 5 t∙ha−1 in favor of \( {\hat{CC}}_{\mathrm{HEMI}} \). Because of these statistics and relationships, we chose \( {\hat{CC}}_{\mathrm{HEMI}} \) for estimating AGB in models with multiple predictor variables.
Top row: Scatterplot for two forms of observed canopy cover (CC) versus observed above-ground biomass (AGB), by vegetation type or land use. CCHEMI is the vertical projection of tree crowns onto the ground, derived from crown area-DBH models; CCHEMI is percentage of the sky blocked by tree crowns measured using a spherical densiometer. Bottom row: Scatterplot for two forms of predicted canopy cover (CC) versus observed above-ground biomass (AGB), by vegetation type or land use, with three models to estimate AGB based on \( {\hat{CC}}_{\mathrm{VERT}} \) or \( {\hat{CC}}_{\mathrm{HEMI}} \)
Models using multiple predictor variables
A clear pattern in reduction of RMSPE can be seen by including more diverse combinations of predictor variable datasets (Table 5). Across models, \( {\hat{CC}}_{\mathrm{HEMI}} \) alone is a better predictor of AGB than models that use only Landsat 8 OLI predictor variables; however, the exponential model that uses Landsat 8 OLI has a slight edge over \( {\hat{CC}}_{\mathrm{HEMI}} \) (RMSPE 45.9 vs. 46.6, respectively). The predictor variable combination of environmental and disturbance data alone and \( {\hat{CC}}_{\mathrm{HEMI}} \) alone are comparable with two of the three combinations that have two predictor variable datasets (Landsat + Env/Dist and \( {\hat{CC}}_{\mathrm{HEMI}} \) + Landsat), with a difference in mean RMSPE% of only three percent. Noticeable improvements in AGB RMSPE% are seen when combining \( {\hat{CC}}_{\mathrm{HEMI}} \) with environmental and disturbance data, as mean RMSPE% drops seven percent. However, the greatest improvement is when performing variable selection using all three predictor variable datasets combined, with a mean RMSPE% of 58 %. Therefore, we performed final variable selection using this combination.
Final variable and model selection
One influential observation, which likely resulted from haze in the remotely-sensed data, was evident across all models. As the influence could be considered uncommon given this dataset, we discarded the observation and continued with the analysis, following the recommendation of Kutner et al. (2005, p.438). Using the final selected variables, we explored use of predictor variable transformations for the sigmoidal and exponential models. By incorporating squared terms in the sigmoidal model for two predictor variables (\( {{\hat{CC}}_{\mathrm{HEMI}}}^2 \) and pH2), we found an improvement of 4 % in terms of RMSE% and an overall better fit in graphs of observed versus estimated values of AGB. The exponential model did not improve by incorporating any transformed predictor variables. However, we found that the fit and validation statistics of the exponential model remained the same after removing one variable (EVI.texture); therefore, we used a subset of five predictor variables for this model. With the GAM, three variables were estimated with a smoothing parameter of 1 (NBR, CTI, bulk density); therefore we fit the model with these variables as parametric terms, as recommended by Wood (2006).
For each of the three models, average RMSE% was less than 60 % and average RMSPE% was less than 65 % (Table 6). The mean prediction difference (MPD) for the sigmoidal model was the highest at 3.2 t∙ha−1; however, mean differences (MD) across all models were less than 1 t∙ha−1. Despite the difference in MPD, the fit and validation statistics for the GAM and the sigmoidal models were otherwise similar, while the exponential model had the worst fit (Fig. 3). Each model had a tendency to under-estimate AGB at higher values, which was most pronounced in the exponential model; however, observations and estimations were relatively balanced along the 1:1 line up to approximately 100 t∙ha−1.
As expected, \( {\hat{CC}}_{\mathrm{HEMI}} \) was selected as a predictor variable in all three models. Vegetation indices were chosen more consistently over raw band values or texture indices among the Landsat 8 OLI predictor variables. The Normalized Difference Moisture Index (NDMI) was chosen in both the GAM and the sigmoidal model, the Normalized Burn Ratio (NBR) was selected as another index in the GAM, while the common Normalized Difference Vegetation Index (NDVI) was selected as the only vegetation index in the exponential model. In the GAM and the sigmoidal model, NDMI was selected in conjunction with topographic or soil variables related to soil moisture (AWC, CTI, D2hydro). Available soil water capacity (AWC) was selected in both the sigmoidal and exponential models. Frequency of late season fire was the only disturbance variable selected, and only in the sigmoidal model.
When each model was applied across the entire study area, AGB ranged from 61.9 to 67.3 t∙ha−1 and total AGB ranged from 64.3 million tonnes to 69.9 million tonnes (Table 7). For comparison, the null model estimate and the simple design-based estimate were both within this same range. All of the model-based methods, including the null model, exhibit confidence intervals that are more precise than the design-based estimate by an order of magnitude. Interestingly, even though the sigmoidal model exhibited the highest mean prediction bias, it had the smallest confidence interval based on bootstrap resampling. Conversely, while the GAM demonstrated the best fit and validation statistics across the three models where variable selection was performed, it had a confidence interval wider than the other models. The sigmoidal model and the exponential model were both more efficient than the null model, while the GAM was less efficient. Since the sigmoidal model had fit and validation statistics similar to the GAM, yet had higher relative efficiency than the GAM, we selected the sigmoidal model to estimate AGB and the corresponding CV for each 20 m × 50 m pixel in the study area.
AGB estimations per pixel using the sigmoidal model ranged from 0 to 390 t∙ha−1, where lower values were more common around areas with higher population and higher values were more common in remote areas in the Luangwa River valley (Fig. 4). Estimates of CV per pixel generally were highest where AGB was the lowest, with a large area of high CV on the eastern boundary and two small valleys in the west half of the study area. The area of high CV on the eastern boundary is dominated by agricultural, nonforest land use; whereas the two small valleys appear to be dominated by grassland. The average CV per pixel across the whole study area was 31 %, with a standard deviation of 14.
Discussion
This research provides encouraging results for using a model-based approach to estimate miombo AGB with National Forest Inventory data. The usefulness of canopy cover as a predictor variable in estimating AGB was clear. Similar results were found by Hall et al. (2006) for boreal forests, by Wu et al. (2013) for tropical woodlands in Sudan, and by González-Roglich and Swenson (2016) for wooded savannas in Argentina. Hall et al. (2006) and González-Roglich and Swenson (2016) used CC as the percentage of the sky blocked by tree crowns over a hemispherical view, whereas Wu et al. used CC as the vertical projection of tree crowns onto the ground. In our case study, CCHEMI (i.e., the hemispherical view of canopy cover) demonstrated a better relationship to AGB for two possible reasons. First, taller trees project more cover in a hemispherical measurement device such as the spherical densiometer (Jennings et al. 1999), and tree height has been shown to be an important covariate in predicting tree-level biomass (Chave et al. 2014). Second, CCVERT (i.e., vertical projection) relies on DBH-crown radius models (Burrows and Strang 1964; Fuller et al. 1997; Isango 2007) to estimate canopy cover per ground plot; these models are outdated and not localized in our study (Halperin et al. 2016). Updating DBH-crown radius models may improve the usefulness of CCVERT as a predictor variable in AGB models for the miombo ecoregion and should be an area of active research. This is especially true for less common vegetation types such as mopane woodlands and riparian forests.
Improvements in AGB estimates were observed when including a diverse range of ecologically meaningful covariates along with \( {\hat{CC}}_{\mathrm{HEMI}} \). In this context, two aspects of moisture stand out. First, soil moisture was characterized by available soil water content (AWC, Hengl et al. 2015) in the sigmoidal and exponential models, or through landscape position via the compound topographic index (CTI) and distance to perennial water in the GAM. Soil moisture has been identified as an underlying factor contributing to miombo vegetation dynamics (Frost 1996; D’Odorico et al. 2007). For example, higher levels of soil moisture have been found under closed canopy miombo woodland compared with open canopy miombo woodland (Campbell et al. 1988).
Second, vegetation moisture was captured through the Normalized Difference Moisture Index (NDMI), an important variable in both the GAM and sigmoidal models. NDMI has been shown to be correlated with tree canopy water content and is competitive with the more well-known Normalized Difference Vegetation Index (NDVI) for a variety of forest mapping purposes (Hunt and Rock 1989; Jin and Sader 2005). This study targeted collection of field and remotely-sensed data for the early dry season, which is considered a prime period for linking these observations in the miombo ecoregion. In this timeframe, grass layers have generally senesced while the deciduous tree canopies have not yet fully shed their leaves (Fuller et al. 1997). As such, NDMI appears to be an important vegetation index for characterizing vegetation moisture in tree canopies, and higher canopy moisture could be related to greater levels of AGB (D’Odorico et al. 2007).
Our study area is in the dry miombo ecozone where rainfall averages less than 1000 mm∙yr−1 (Timberlake and Chidumayo 2011). Comparing our results with similar studies in the same ecozone, Næsset et al. (2016) estimated mean AGB at 51.3 t∙ha−1, Mauya et al. (2015) at 65.8 t∙ha−1, Ryan et al. (2011) at 59.4 t∙ha−1, and Ribeiro et al. (2008) at 70 t∙ha−1, while Carreiras et al. (2013) estimated AGB at 25.7 t∙ha−1. However, Carreiras et al. only studied areas with canopy cover < 50 %. The review by Frost (1996) provided an AGB estimate of 55 t∙ha−1for dry miombo of Zambia and Zimbabwe, and in the most recent FAO Forest Resources Assessment for Zambia, the national estimate of AGB was 84 t∙ha−1 (FAO 2014). Across models, our AGB estimates ranged from 61.9 to 67.3 t∙ha−1, where the best performing model and the null model estimated 62.1 and 62.8 t∙ha−1, respectively. Clearly, our estimates of AGB for a study area in the Zambian dry miombo ecozone are in line with other studies.
Comparing accuracy statistics with other published results is challenging, given the number of studies using similar data sources in the same ecozone, as well as the use of a wide variety of statistics. Therefore, a comparison of similar studies estimating AGB in other miombo ecozones or dry forest ecoregions is necessary. Some researchers presented model fit statistics such as R 2 (Wu et al. 2013; Kashindye et al. 2013), while others emphasized RMSE (Mitchard et al. 2013; Solberg et al. 2015). The latter two estimated RMSE at 12.8 t∙ha−1 (22 % CV) in Mozambique and 40.3 t∙ha−1 (78 % CV) in Tanzania, respectively. However, fit statistics are known to be optimistic when concerning prediction (Guisan and Zimmermann 2000; Packalén et al. 2012). Using resampling methods to estimate prediction error for AGB in Mozambique, Ryan et al. (2012) and Carreiras et al. (2013) reported RMSPE of 19.6 t∙ha−1 and 5.0 t∙ha−1, respectively, where the latter reported a mean CV of 25 %. In a study site in Tanzania with similar vegetation and rainfall to our study area, Mauya et al. (2015) and Næsset et al. (2016) reported RMSPE% of 47 and 62 %, respectively. These latter studies all employed active sensor (radar or ALS) imagery as the single source for predictor variables.
Research on the use of optical sensor satellite imagery for estimating AGB in the miombo ecoregion is more limited. Næsset et al. (2016) used a linear ordinary least squares model and a stepwise predictor variable selection method. As potential predictor variables, these authors included canopy cover predictions and canopy cover gain or loss from Hansen et al. (2013), raw band values from Landsat 7 ETM+ and Landsat 8 OLI, NDVI calculated from the Landsat data, and square and square root transformations for all of these predictor variables. Based on their stepwise predictor variable selection procedure, squared canopy cover was the only predictor variable chosen. While their finding helps to corroborate our result of canopy cover and squared canopy cover in the sigmoidal model, their model resulted in poor AGB prediction with an RMSPE% of 87 %. It is possible that their model using Landsat data was disadvantaged by two aspects. First, they did not include other commonly used vegetation indices (i.e., NDMI, NBR) or texture variables (i.e. standard deviation in a 3 × 3 window around the ground plot), which we found to be useful in our study, as have others (e.g., Lu et al. 2014). Second, the Landsat data that Næsset et al. used differed by at least a year from the time of their ground plot measurements.
Two studies that estimated miombo tree volume (m3∙ha−1) using Landsat also provide a useful comparison. Employing cross-validation to compute RMSPE%, Pereira (2006) reported 48 % in Mozambique using Landsat and k-Nearest Neighbor imputation, while Gara et al. (2015) averaged 62 % using Landsat and nonlinear models. Our chosen model (sigmoidal) has estimates of RMSE% at 47 and RMSPE% at 58 %, indicating that our fit and validation statistics are comparable to similar studies using large-scale inventories in the miombo ecoregion using optical remotely-sensed data (Pereira 2006) and active remote sensing data such as ALS or radar (Solberg et al. 2015; Mauya et al. 2015; Næsset et al. 2016). However, more research is needed to compare AGB confidence intervals to other studies because application of a model-based approach appears relatively uncommon in the miombo ecoregion.
The use of GAMs and nonlinear models, such as the sigmoidal and exponential models in this study, for predicting AGB has shown promise in a number of studies in different biomes (e.g., Moisen and Frescino 2002; Hall et al. 2006; Anaya et al. 2009; McRoberts et al. 2015; Kattenborn et al. 2015). However, it appears that most approaches to estimate and map AGB in the miombo ecoregion used linear ordinary least squares (Ryan et al. 2012; Kashindye et al. 2013; Mitchard et al. 2013; Solberg et al. 2015; Næsset et al. 2016), despite the nonlinear relationships between forest attributes and remotely-sensed data (Sedano et al. 2008; Banskota et al. 2014; Lu et al. 2016 ; Gara et al. 2015). We found GAM’s to perform as well as nonlinear models with respect to fit and validation statistics; however, when applied to the population of predictor data the bootstrap confidence interval was less efficient than the null model.
We protected against overfitting of the GAM by using cross-validation in the variable selection process (Guisan and Zimmermann 2000; Hawkins 2004; Packalén et al. 2012); however, the smoothing splines of GAM’s may perform in an unpredictable manner when applied to population-level data which includes ranges of values not within the observation data (Guisan et al. 2002; Venables and Dichmont 2004). Because of this, nonlinear models may be preferred for several reasons. First, the sigmoidal and exponential models in this study are asymptotic, thereby preventing both negative and unreasonably high estimations (McRoberts et al. 2015). Second, nonlinear models are able to achieve more stable extrapolation at the ends of, and beyond, the ranges of the values of the predictor variables (Venables and Dichmont 2004). Third, judicious application of transformations may improve performance if there appears to be a lack of fit in nonlinear models.
One issue with the nonlinear models we used in this study is estimating the asymptote. We initially investigated a flexible asymptote (α), with α estimated in the model fitting process. For our dataset, α was often estimated at unrealistically high values or the nonlinear models would not converge, both of which may indicate that the data does not support estimating a biologically meaningful asymptote. We further investigated estimating α as a function of the predictor variables in the models (i.e., α0 + α1 x i ); however, no improvements in model fit or biologically meaningful values of α were noted. In this context, Burkhart and Tomé (2012 p. 123) stated that if an asymptote cannot be realistically fit with the data at hand, then expert judgement may be employed to fix the asymptote at a reasonable value and continue with model fitting. AGB t∙ha−1 for most vegetation types in the dry miombo ecozone tend to be less than 100 t∙ha−1 (Frost 1996), although upper limits generally appear to be understudied. Mauya et al. (2015) found an upper limit of 350 t∙ha−1 in dry miombo woodland vegetation type for a study site in southeast Tanzania, while the riparian forest vegetation type may approach 500 t∙ha−1 (Chidumayo personal communication 2016). Future modeling efforts should assess practical asymptotic limits for AGB in all miombo vegetation types.
While this study made progress in terms of ecologically meaningful variable selection and highlighted model advantages and disadvantages, improvements may be realized in several aspects. First, we found that canopy cover as the percentage of the sky blocked by tree crowns over a hemispherical view (\( {\hat{CC}}_{\mathrm{HEMI}} \)) was a better predictor for AGB than canopy cover as the vertical projection of tree crowns onto the ground (\( {\hat{CC}}_{\mathrm{VERT}} \)). However, values of CCVERT, used to predict \( {\hat{CC}}_{\mathrm{VERT}} \), were derived from DBH-crown radius models (Burrows and Strang 1964; Fuller et al. 1997; Isango 2007) that should be updated for conditions in Zambia (Halperin et al. 2016). Once updated, \( {\hat{CC}}_{\mathrm{VERT}} \) may prove to be as useful as \( {\hat{CC}}_{\mathrm{HEMI}} \) as a predictor of AGB. Second, the highest percent CV pixel values occurred where AGB was predicted to be the lowest, likely because there were very few ground plots with no trees. To accommodate such under-sampled areas, off-grid ground plots could be included along with the NFI ground plots in implementation of the model-based methods. Model-based methods are not restricted to one single sample design as long as the same variable of interest (i.e., AGB) is being sampled according to the same definitions; the main consideration is that sample selection is required to be uninformative in terms of Y (Gregoire 1998).
Third, active sensor remotely-sensed data allow tree heights to be estimated and height is known be correlated with tree-level (Chave et al. 2014) and plot-level biomass (Wulder et al. 2012). ALS data is a proven source of height estimates, yet remains operationally out-of-reach for many countries (Ribeiro et al. 2012). Testing of this source of predictor variables for use in operational monitoring programs is just now underway in the miombo ecoregion (Mauya et al. 2015; Næsset et al. 2016). Additionally, significant challenges remain in using more readily available radar data (Sinha et al. 2015; Solberg et al. 2015). However, space borne laser scanning systems are under development (Moussavi et al. 2014; GOFC-GOLD 2015) and may prove useful in future applications.
Fourth, a model-assisted approach may lend itself to use of the GAM, balancing the variability in population-level estimation with more conservative inference that takes advantage of the design-based properties of the sample (Opsomer et al. 2007; Næsset et al. 2016). Fifth, there is significant opportunity to explore model-assisted or model-based approaches in a small area estimation context for improving both estimates and inference at spatial scales where traditional design-based approaches are not possible (Magnussen et al. 2014). Lastly, if a map of AGB is a requirement, then use of georeferenced predictor variables is required. However, if a map of AGB is not required, then our null model would provide a viable alternative that could also be applied within a small area estimation framework.
Conclusion
Estimating the amount and distribution of AGB is crucial for improving land management planning and for engaging in international discussions on REDD+. Our understanding of AGB is enhanced by including ecologically meaningful predictor variables in a model-based approach to estimation. This study generated several insights for improving miombo AGB estimation using NFI data. First, the biophysical relationship between AGB and CC can be exploited to improve AGB estimation. Second, model-based methods improve precision of AGB estimates by an order of magnitude over simple design-based estimation. Third, if a map of AGB is not required, estimating the confidence interval of total AGB using bootstrap resampling for a null model is a viable alternative to the traditional design-based confidence interval. Fourth, if a map is required, expensive predictor variables such as those derived from ALS, are not necessarily required as there is freely available spatial data (i.e., Landsat, Digital Elevation Model, soils, etc.) which can be used as predictor variables in the model-based framework. Fifth, a genetic algorithm provides an efficient way to perform predictor variable selection. Lastly, future approaches to improve our understanding of AGB distribution in the miombo ecoregion should include small area estimation.
Abbreviations
AGB, above-ground biomass; ALS, airborne laser scanning; CC, canopy cover; CV, Coefficient of Variation; GA, genetic algorithm; GAM, generalized additive model; NFI, National Forest Inventory; OLI, Operational Land Imager; REDD+, Reducing Emissions from Deforestation and forest Degradation; RMSE, root mean square error; RMSPE, root mean square prediction error; t∙ha−1, tonnes per hectare
References
Ahrends A, Burgess ND, Milledge SA, Bulling MT, Fisher B, Smart JC, Clarke G, Mhorok BE, Lewis SL (2010) Predictable waves of sequential forest degradation and biodiversity loss spreading from an African city. Proc Natl Acad Sci 107(33):14556–14561
Anaya JA, Chuvieco E, Palacios-Orueta A (2009) Above-ground biomass assessment in Colombia: a remote sensing approach. For Ecol Manage 257(4):1237–1246
Banskota A, Kayastha N, Falkowski MJ, Wulder MA, Froese RE, White JC (2014) Forest monitoring using Landsat time series data: a review. Can J Remote Sens 40(5):362–384
Bater CW, Coops NC, Gergel SE, LeMay V, Collins D (2009) Estimation of standing dead tree class distributions in northwest coastal forests using lidar remote sensing. Can J Forest Res 39(6):1080–1091
Boschetti L, Roy D, Hoffmann AA (2013) MODIS Collection 5.1 Burned Area Product-MCD45. User’s Guide. Ver, 3.0.1. University of Maryland, College Park, Maryland, USA
Brink AB, Bodart C, Brodsky L, Defourney P, Ernst C, Donney F, Lupi A, Tuckova K (2014) Anthropogenic pressure in East Africa—Monitoring 20 years of land cover changes by means of medium resolution satellite data. Int J Appl Earth Obs Geoinf 28:60–69
Burkhart HE, Tomé M (2012) Modeling forest trees and stands. Springer Science & Business Media, New York. p 461
Burrows PM, Strang RM (1964) The relation of crown and basal diameters in Rhodesian Brachystegia woodland. The Commonwealth Forestry Review Vol. 43, No. 4 (118), pp. 331-333
Cabral AIR, Vasconcelos MJ, Oom D, Sardinha R (2010) Spatial dynamics and quantification of deforestation in the central-plateau woodlands of Angola (1990–2009). Appl Geogr 31(3):1185–1193
Campbell BM, Swift MJ, Hatton J, Frost PGH (1988) Small-scale vegetation pattern and nutrient cycling in miombo woodland. In: Verhoeven JTA, Heil GW, Werger M (eds) Vegetation structure in relation to carbon and nutrient economy. SPB Academic Publishing, The Hague, pp 69–85
Carreiras J, Melo JB, Vasconcelos MJ (2013) Estimating the above-ground biomass in miombo savanna woodlands (Mozambique, East Africa) using L-band synthetic aperture radar data. Remote Sens 5(4):1524–1548
Chave J, Réjou‐Méchain M, Búrquez A, Chidumayo E, Colgan MS, Delitti WBC, Duque A, Eid T, Fearnside PM, Goodman RC, Henry M, Martinez-yrizar A, Mugasha WA, Muller Landau HC, Mencuccini M, Nelson BW, Ngomanda A, Nogueira EM, Ortiz-malavassi E, Pelissier R, Ploton P, Ryan CM, Saldarriaga J, Vieilleden G (2014) Improved allometric models to estimate the above-ground biomass of tropical trees. Glob Chang Biol 20(10):3177–3190
Chidumayo EN (1988) A re-assessment of effects of fire on miombo regeneration in the Zambian Copperbelt. J Trop Ecol 4(4):361–372
Chidumayo E, Gambiza J, Grundy I (1996) Managing miombo woodlands. In: Campbell B (ed) The Miombo in transition: woodlands and welfare in Africa. Center for International Forestry Research, Bogor, pp 175–193, P 266
Chidumayo EN (2012) Assessment of existing models for biomass and volume calculations for Zambia. Report prepared for FAO-Zambia and the Integrated Land Use Assessment (ILUA) Phase 2 Project. Food and Agriculture Organization of the United Nations, Rome
Cochran WG (1977) Sampling techniques, 3rd edn. Wiley, New York, p 428
D’Odorico P, Caylor K, Okin GS, Scanlon TM (2007) On soil moisture–vegetation feedbacks and their possible effects on the dynamics of dryland ecosystems. J Geophys Res Biogeosci (2005–2012) 112:G4
Efron B, Tibshirani R (1986) Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat Sci 1(1):54–75
ESRI (2012) ArcGIS desktop: release 10.1. Environmental Systems Research Institute, Redlands
FAO (2014) Zambia - Global Forest Resources Assessment 2015 – Country Report. Rome. p 76
Faßnacht FE, Hartig F, Latifi H, Berger C, Hernández J, Corvalán P, Koch B (2014) Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens Environ 154:102–114
Frost P (1996) The ecology of miombo woodlands. In: Campbell B (ed) The Miombo in transition: woodlands and welfare in Africa (pp 11–57). Center for International Forestry Research, Bogor, p 266
Fuller DO, Prince SD, Astle WL (1997) The influence of canopy strata on remotely sensed observations of savanna-woodlands. Int J Remote Sens 18(14):2985–3009
Gara TW, Murwira A, Ndaimani H, Chivhenge E, Hatendi CM (2015) Indigenous forest wood volume estimation in a dry savanna, Zimbabwe: exploring the performance of high-and-medium spatial resolution multispectral sensors. Transactions of the Royal Society of South Africa, Vol. 70, No. 3, 285–293
Garcia-Gutierrez J, Gonzalez-Ferreiro E, Riquelme-Santos JC, Miranda D, Dieguez-Aranda U, Navarro-Cerrillo RM (2014) Evolutionary feature selection to estimate forest stand variables using LiDAR. Int J Appl Earth Obs Geoinf 26:119–131
GOFC-GOLD (2015) A sourcebook of methods and procedures for monitoring and reporting anthropogenic greenhouse gas emissions and removals associated with deforestation, gains and losses of carbon stocks in forests remaining forests, and forestation. GOFC-GOLD Report version COP21-1. GOFC-GOLD Land Cover Project Office, Wageningen University, The Netherlands
González-Roglich M, Swenson JJ, Jobbágy EG, Jackson RB (2014) Shifting carbon pools along a plant cover gradient in woody encroached savannas of central Argentina. Forest Ecol Manag 331:71–78
González-Roglich M, Swenson JJ (2016) Tree cover and carbon mapping of Argentine savannas: scaling from field to region. Remote Sens Environ 172:139–147
GRZ (Government of the Republic of Zambia) (1968) The Republic of Zambia – National rainfall map, 1:3,000,000. Government Printer, Lusaka. Accessed online at http://eusoils.jrc.ec.europa.eu/esdb_archive/eudasm/africa/lists/czm.htm on 11 April, 2014
GRZ (Government of the Republic of Zambia) (1976) Vegetation map, 1:500,000. Survey Department, Lusaka, Sheets 1 – 9
GRZ (Government of the Republic of Zambia) (1986) The Republic of Zambia – National soil map, 1:3,000,000. Government Printer, Lusaka. Accessed online at http://esdac.jrc.ec.europa.eu/resource-type/national-soil-maps-eudasm on 21 July 2016
GRZ (Government of the Republic of Zambia) (2014) Integrated Land Use Assessment Phase 2 - Zambia Biophysical Field Manual, version March 25, 2014. Forestry Department. Ministry of Lands, Natural Resources and Environmental Protection in cooperation with Food and Agriculture Organization (FAO), Lusaka
Gregoire TG (1998) Design-based and model-based inference in survey sampling: appreciating the difference. Can J Forest Res 28(10):1429–1447
Guisan A, Zimmermann NE (2000) Predictive habitat distribution models in ecology. Ecol Model 135(2):147–186
Guisan A, Edwards TC, Hastie T (2002) Generalized linear and generalized additive models in studies of species distributions: setting the scene. Ecol Model 157(2):89–100
Gumbo DJ, Mumba KY, Kaliwile MM, Moombe KB, Mfuni TI (2016) Agrarian changes in the Nyimba District of Zambia. In: Deakin L, Kshatriya M, Sunderland T (eds) Agrarian change in tropical landscapes. Center for International Forestry Research, Bogor, pp 234–268
Halperin J, LeMay V, Coops N, Verchot L, Marshall P, Lochhead K (2016) Canopy cover estimation in miombo woodlands of Zambia: comparison of Landsat 8 OLI versus RapidEye imagery using parametric, nonparametric, and semiparametric methods. Remote Sens Environ 179:170–182
Hall RJ, Skakun RS, Arsenault EJ, Case BS (2006) Modeling forest stand structure attributes using Landsat ETM+ data: application to mapping of aboveground biomass and stand volume. Forest Ecol Manag 225(1):378–390
Hansen MC, DeFries RS, Townshend JRG, Marufu L, Sohlberg R (2002) Development of a MODIS tree cover validation data set for Western Province, Zambia. Remote Sens Environ 83(1):320–335
Hansen MC, Potapov PV, Moore R, Hancher M, Turubanova SA, Tyukavina A, Thau D, Stehman SV, Goetz SJ, Loveland TR, Kommareddy A (2013) High-resolution global maps of 21st-century forest cover change. Science 342(6160):850–853
Hawkins DM (2004) The problem of overfitting. J Chem Inf Comput Sci 44(1):1–12
Helmer EH, Brandeis TJ, Lugo AE, Kennaway T (2008) Factors influencing spatial pattern in tropical forest clearance and stand age: implications for carbon storage and species diversity. J Geophys Res 113:G02S04. doi:10.1029/2007JG000568
Hengl T, Heuvelink GB, Kempen B, Leenaars JG, Walsh MG, Shepherd KD, Sila A, MacMillan RA, de Jesus JM, Tamene L, Tondoh JE (2015) Mapping soil properties of Africa at 250 m resolution: random forests significantly improve current predictions. PLoS One 10(6):e0125814
Hijmans RJ, van Etten J, Mattiuzzi M, Sumner M, Greenberg J, Lamigueiro O, Bevan A, Racine E, Shortridge A (2015) Raster: geographic analysis and modeling with raster data. R package version 2., pp 3–41, http://cran.r-project.org/web/packages/raster/index.html
Holmgren PLG, Marklund F (2007) National forest monitoring systems: purposes, options and status. In: Freer-Smith PH, Broadmeadow MS, Lynch JM (eds) Forestry and climate change. CAB International, UK, pp 163–173
Hunt ER, Rock BN (1989) Detection of changes in leaf water content using near-and middle-infrared reflectances. Remote Sens Environ 30(1):43–54
Isango JA (2007) Stand structure and tree species composition of Tanzania miombo woodlands: a case study from miombo woodlands of community based forest management in Iringa District. Working Papers of the Finnish Forest Research Institute 50:43–56
Jennings SB, Brown ND, Sheil D (1999) Assessing forest canopies and understory illumination: canopy closure, canopy cover and other measures. Forestry 72(1):59–74
Jin S, Sader SA (2005) Comparison of time series tasseled cap wetness and the normalized difference moisture index in detecting forest disturbances. Remote Sens Environ 94(3):364–372
Justice CO, Vermote E, Townshend JRG, DeFries R, Roy DP, Hall DK, Salomonson VV, Privette JL, Riggs G, Strahler A, Lucht W, Myneni RB, Knyazikhin Y, Running SW, Nemani RR, Zhengming W, Huete AR, Van Leeuwen W, Wolfe RE, Giglio L, Muller J-P, Lewis P, Barnsley MJ (1998) The Moderate Resolution Imaging Spectroradiometer (MODIS): Land remote sensing for global change research. Geosci Remote Sens IEEE Trans 36(4):1228–1249
Kashindye A, Mtalo E, Mpanda MM, Liwa E, Giliba R (2013) Multi-temporal assessment of forest cover, stocking parameters and above-ground tree biomass dynamics in miombo woodlands of Tanzania. Afr J Environ Sci Technol 7(7):611–623
Kaufman YJ, Tanre D (1992) Atmospherically resistant vegetation index (ARVI) for EOS-MODIS. Geosci Remote Sens IEEE Trans 30(2):261–270
Kattenborn T, Maack J, Faßnacht F, Enßle F, Ermert J, Koch B (2015) Mapping forest biomass from space – Fusion of hyperspectral EO1-hyperion data and Tandem-X and WorldView-2 canopy height models. Intl Appl Earth Observ Geoinform 35:359–367
Key CH, Benson NC (2006) Landscape assessment: remote sensing of severity, the Normalized Burn Ratio. In: Lutes DC (ed) FIREMON: fire effects monitoring and inventory system. RMRS-GTR-164-CD. USDA, Forest Service, Rocky Mountain Research Station, Fort Collins
Kutner MH, Nachtsheim CJ, Neter J, Li W (2005) Applied linear statistical models, 5th ed. McGraw Hill, New York
Lefsky MA, Cohen WB, Harding DJ, Parker GG, Acker SA, Gower ST (2002) Lidar remote sensing of above‐ground biomass in three biomes. Glob Ecol Biogeogr 11(5):393–399
Lefsky MA, Cohen WB (2003) Selection of remotely sensed data. In: Wulder M, Franklin SE (eds) Remote sensing of forest environments: concepts and case studies. Springer Science & Business Media, Springer US, pp 535–546
Lu D, Chen Q, Wang G, Liu L, Li G, Moran E (2016) A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. International Journal of Digital Earth, 9(1), 63-105. doi:10.1080/17538947.2014.990526
Makhado RA, Mapaure I, Potgieter MJ, Luus-Powell WJ, Saidi AT (2014) Factors influencing the adaptation and distribution of Colophospermum mopane in southern Africa’s mopane savannas – a review. Bothalia 44(1):9
Magnussen S, Mandallaz D, Breidenbach J, Lanz A, Ginzler C (2014) National forest inventories in the service of small area estimation of stem volume. Can J Forest Res 44(9):1079–1090
Masek JG, Vermote EF, Saleous NE, Wolfe R, Hall FG, Huemmrich KF, Gao F, Kutler J, Lim TK (2006) A Landsat surface reflectance dataset for North America, 1990–2000. Geosci Remote Sens Lett IEEE 3(1):68–72
Mauya EW, Ene LT, Bollandsås OM, Gobakken T, Næsset E, Malimbwi RE, Zahabu E (2015) Modelling aboveground forest biomass using airborne laser scanner data in the miombo woodlands of Tanzania. Carbon Balance Manage 10(1):1–16
Mayes MT, Mustard JF, Melillo JM (2015) Forest cover change in miombo woodlands: modeling land cover of African dry tropical forests with linear spectral mixture analysis. Remote Sens Environ 165:203–215
McRoberts RE, Tomppo EO, Næsset E (2010) Advances and emerging issues in national forest inventories. Scand J Forest Res 25(4):368–381
McRoberts RE, Magnussen S, Tomppo EO, Chirici G (2011) Parametric, bootstrap, and jackknife variance estimators for the k-Nearest Neighbors technique with illustrations using forest inventory and satellite image data. Remote Sens Environ 115(12):3165–3174
McRoberts RE, Næsset E, Gobakken T (2015) The effects of temporal differences between map and ground data on map-assisted estimates of forest area and biomass. Annals of Forest Science. doi: 10.1007/s13595-015-0485-6
Mitchard ET, Meir P, Ryan CM, Woollen ES, Williams M, Goodman LE, Mucavele J, Watts P, Woodhouse I, Saatchi SS (2013) A novel application of satellite radar data: measuring carbon sequestration and detecting degradation in a community forestry project in Mozambique. Plant Ecol Divers 6(1):159–170
Moisen GG, Frescino TS (2002) Comparing five modelling techniques for predicting forest characteristics. Ecol Model 157(2):209–225
Moisen GG, Freeman EA, Blackard JA, Frescino TS, Zimmermann NE, Edwards TC Jr (2006) Predicting tree species presence and basal area in Utah: a comparison of stochastic gradient boosting, generalized additive models, and tree-based methods. Ecol Model 199(2):176–187
Moussavi MS, Abdalati W, Scambos T, Neuenschwander A (2014) Applicability of an automatic surface detection approach to micro-pulse photon-counting LiDAR altimetry data: implications for canopy height retrieval from future ICESat-2 data. Int J Remote Sens 35(13):5263–5279
Næsset E, Ørka HO, Solberg S, Bollandsås OM, Hansen EH, Mauya E, Zahabu E, Malimbwi R, Chamuya N, Olsson H, Gobakken T (2016) Mapping and estimating forest area and aboveground biomass in miombo woodlands in Tanzania using data from airborne laser scanning, TanDEM-X, RapidEye, and global forest maps: a comparison of estimated precision. Remote Sens Environ 175:282–300
Opsomer JD, Breidt FJ, Moisen GG, Kauermann G (2007) Model-assisted estimation of forest resources with generalized additive models. J Am Stat Assoc 102(478):400–409
Packalén P, Temesgen H, Maltamo M (2012) Variable selection strategies for nearest neighbor imputation methods used in remote sensing based forest inventory. Can J Remote Sens 38(5):557–569
Pereira CR (2006) Estimating and mapping forest inventory variables using the k-NN method: Mocuba District case study – Mozambique. PhD dissertation, Universita Tuscia, Viterbo, Italy. p 86
Pflugmacher D, Cohen WB, Kennedy RE, Yang Z (2014) Using Landsat-derived disturbance and recovery history and lidar to map forest biomass dynamics. Remote Sens Environ 151:124–137
Powell SL, Cohen WB, Healey SP, Kennedy RE, Moisen GG, Pierce KB, Ohmann JL (2010) Quantification of live aboveground forest biomass dynamics with Landsat time-series and field inventory data: A comparison of empirical modeling approaches. Remote Sens Environ 114(5):1053–1068
R Development Core Team (2015) R: A Language and Environment for Statistical Computing, version 3.2.2 (August 2015). R Foundation for Statistical Computing, Vienna, http://www.R-project.org
Ribeiro NS, Saatchi SS, Shugart HH, Washington‐Allen RA (2008) Aboveground biomass and leaf area index (LAI) mapping for Niassa Reserve, northern Mozambique. J Geophys Res Biogeosci (2005–2012) 113:G3
Ribeiro N, Cumbana M, Mamugy F, Chaúque A (2012) Remote sensing of biomass in the miombo woodlands of southern Africa: opportunities and limitations for research. In: Fatoyinbo (Ed.) Remote Sensing of Biomass - Principles and Applications, p 77–98. ISBN: 978-953-51-0313-4, InTech, Rijeka, Croatia
Rouse JW, Jr Haas RH, Schell JA, Deering DW (1974) Monitoring vegetation systems in the Great Plains with ERTS. In: Freden SC, Mercanti EP, Becker MA (eds) Technical Presentations Sec. A, vol 1. NASA Science and Technology Information Office, Washington, pp 309–317
Ryan CM, Williams M (2011) How does fire intensity and frequency affect miombo woodland tree populations and biomass? Ecol Appl 21(1):48–60
Ryan CM, Williams M, Grace J (2011) Above‐and belowground carbon stocks in a miombo woodland landscape of Mozambique. Biotropica 43(4):423–432
Ryan CM, Hill T, Woollen E, Ghee C, Mitchard E, Cassells G, Grace J, Woodhouse IH, Williams M (2012) Quantifying small‐scale deforestation and forest degradation in African woodlands using radar imagery. Glob Change Biol 18(1):243–257
Samimi C, Kraus T (2004) Biomass estimation using Landsat-TM and-ETM+. Towards a regional model for Southern Africa? GeoJournal 59(3):177–187
Sankaran M, Ratnam J, Hanan N (2008) Woody cover in African savannas: the role of resources, fire and herbivory. Glob Ecol Biogeogr 17(2):236–245
Sawe TC, Munishi PK, Maliondo SM (2014) Woodlands degradation in the Southern Highlands miombo of Tanzania: implications on conservation and carbon stocks. Int J Biodiv Conserv 6(3):230–237
Scrucca L (2013) GA: a package for Genetic Algorithms in R. J Stat Softw 53(4):1–37
Sedano F, Gómez D, Gong P, Biging GS (2008) Tree density estimation in a tropical woodland ecosystem with multiangular MISR and MODIS data. Remote Sens Environ 112(5):2523–2537
Shearman P, Bryan J, Laurance WF (2012) Are we approaching ‘peak timber’ in the tropics? Biol Conserv 151(1):17–21
Sinha S, Jeganathan C, Sharma LK, Nathawat MS (2015) A review of radar remote sensing for biomass estimation. Int J Environm Sci Techn 12(5):1779–1792
Skutsch MM, Ba L (2010) Crediting carbon in dry forests: the potential for community forest management in West Africa. Forest Policy Econ 12(4):264–270
Solberg S, Gizachew B, Næsset E, Gobakken T, Bollandsås OM, Mauya EW, Olsson H, Malimbwi R, Zahabu E (2015) Monitoring forest carbon in a Tanzanian woodland using interferometric SAR: a novel methodology for REDD+. Carbon Balance Manage 10(1):1–14
Ståhl G, Saarela S, Schnell S, Holm S, Breidenbach J, Healey SP, Patterson PL, Magnussen S, Næsset E, McRoberts RE, Gregoire TG (2016) Use of models in large-area forest surveys: comparing model-assisted, model-based and hybrid estimation. Forest Ecosyst 3(1):1
Stringer LC, Dougill AJ, Thomas AD, Spracklen DV, Chesterman S, Speranza CI, Rueff H, Riddell M, Williams M, Beedy T, Abson DJ, Klintenberg P, Syampungani S, Powell P, Palmer AR, Seely MK, Mkwambisi DD, Falcao M, Sitoe A, Ross S, Kopolo G (2012) Challenges and opportunities in linking carbon sequestration, livelihoods and ecosystem service provision in drylands. Environ Sci Policy 19:121–135
Suberu MY, Bashir N, Mustafa MW (2014) Over use of wood-based bioenergy in selected sub-Saharan Africa countries: review of unconstructive challenges and suggestions. J Clean Prod. doi:10.1016/j.jclepro.2014.04.014
Suganuma H, Abe Y, Taniguchi M, Tanouchi H, Utsugi H, Kojima T, Yamada K (2006) Stand biomass estimation method by canopy coverage for application to remote sensing in an arid area of Western Australia. Forest Ecol Manage 222(1):75–87
Timberlake JR (1995) Colophospermum mopane: annotated bibliography and review. The Zimbabwe Bulletin of Forestry Research, No. 11. ISBN: 0-7974-1420-7., p 49
Timberlake J, Chidumayo EN (2011) Miombo ecoregion vision report. Occasional Publications in Biodiversity No. 20 Biodiversity Foundation for Africa, Bulawayo
Timilsina N, Staudhammer CL (2012) Individual tree mortality model for slash pine in Florida: a mixed modeling approach. South J Appl Forest 36(4):211–219
Tiwari AK, Singh JS (1984) Mapping forest biomass in India through aerial photographs and nondestructive field sampling. Appl Geogr 4(2):151–165
Tomppo E, Halme M (2004) Using coarse scale forest variables as ancillary information and weighting of variables in k-NN estimation: a genetic algorithm approach. Remote Sens Environ 92(1):1–20
USGS (United States Geological Survey) (2004) Shuttle Radar Topography Mission, 1 Arc Second SRTM s14e29, s14e30, s14e31, s15e29, s15e30, s15e31, s16e29, s16e30, s16e31, Unfilled Unfinished 2.0. Global Land Cover Facility, University of Maryland, College Park, February 2000
Venables WN, Dichmont CM (2004) GLMs, GAMs and GLMMs: an overview of theory for applications in fisheries research. Fisheries Res 70(2):319–337
Wang Y, LeMay VM, Baker TG (2007) Modelling and prediction of dominant height and site index of Eucalyptus globulus plantations using a nonlinear mixed-effects model approach. Can J Forest Res 37(8):1390–1403
Wood SN (2006) Generalized Additive Models: an introduction with R. Chapman & Hall/CRC Press, Chicago
Wood SN (2015) Reference manual for mgcv: Mixed GAM Computation Vehicle with GCV/AIC/REML smoothness estimation, v 1.8-9 (30 Oct 2015). Available at: http://cran.r-project.org/web/packages/mgcv/index.html
Wu W, De Pauw E, Helldén U (2013) Assessing woody biomass in African tropical savannahs by multiscale remote sensing. Int J Remote Sens 34(13):4525–4549
Wulder MA, White JC, Nelson RF, Næsset E, Ørka HO, Coops NC, Hilker T, Bater CW, Gobakken T (2012) Lidar sampling for large-area forest characterization: a review. Remote Sens Environ 121:196–209
Acknowledgements
The authors wish to thank Dr. Nicholas Coops, Dr. Davison Gumbo, Mr. Kaala Moombe, Mr. Joel Lwambo, Mr. Martin Lyambai, Mr. Andrew Goods Nkoma, Chief Nyalugwe, Chief Ndake, Chieftaness Mwape, Chief Luembe, Mrs. Bertha Kauseni, Ms. Rhoda Chiluba, Mr. Shadreck Ngoma, Mr. Sylvester Siame, Mr. Geoffrey Tebuho, Mr. Susiku Muyapekwa, Mr. Smart Lungu, Mr. Saule Lungu, Mr. Moses Ngulube, Dr. Julian Fox, Mr. Abel Siampale, and the residents of Nyimba District, Zambia. We also wish to thank two anonymous reviewers for their helpful comments and suggestions.
Funding
Funding for the field work in this study was provided by the United States Agency for International Development under grant number BFS-G-11-00002 to the Center for International Forestry Research, entitled the Nyimba Forest Project. Funding for the lead author to conduct analysis has been provided by The University of British Columbia.
Authors’ contributions
JH designed the study, led the field work, analyzed the data, and wrote the manuscript. VL supervised the study design, analysis, and initial manuscript drafts. EN, LV, and PM provided essential suggestions and advice throughout the process. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Halperin, J., LeMay, V., Chidumayo, E. et al. Model-based estimation of above-ground biomass in the miombo ecoregion of Zambia. For. Ecosyst. 3, 14 (2016). https://doi.org/10.1186/s40663-016-0077-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40663-016-0077-4