Abstract
Background
Many tree species in tropical forests have distributions tracking local ridge-slope-valley topography. Previous work in a 50-ha plot in Korup National Park, Cameroon, demonstrated that 272 species, or 63% of those tested, were significantly associated with topography.
Methods
We used two censuses of 329,000 trees ≥1 cm dbh to examine demographic variation at this site that would account for those observed habitat preferences. We tested two predictions. First, within a given topographic habitat, species specializing on that habitat (‘residents’) should outperform species that are specialists of other habitats (‘foreigners’). Second, across different topographic habitats, species should perform best in the habitat on which they specialize (‘home’) compared to other habitats (‘away’). Species’ performance was estimated using growth and mortality rates.
Results
In hierarchical models with species identity as a random effect, we found no evidence of a demographic advantage to resident species. Indeed, growth rates were most often higher for foreign species. Similarly, comparisons of species on their home vs. away habitats revealed no sign of a performance advantage on the home habitat.
Conclusions
We reject the hypothesis that species distributions along a ridge-valley catena at Korup are caused by species differences in trees ≥1 cm dbh. Since there must be a demographic cause for habitat specialization, we offer three alternatives. First, the demographic advantage specialists have at home occurs at the reproductive or seedling stage, in sizes smaller than we census in the forest plot. Second, species may have higher performance on their preferred habitat when density is low, but when population builds up, there are negative density-dependent feedbacks that reduce performance. Third, demographic filtering may be produced by extreme environmental conditions that we did not observe during the census interval.
Similar content being viewed by others
Background
A common feature of species-rich forests is high beta diversity resulting from turnover in tree species composition across habitat types (Shmida and Wilson [1985]; Condit et al. [2002]; Paoli et al. [2006]). Turnover results from differences in how species respond to climate and soil gradients. At a local scale, within a few hundred meters, it is common to observe species turnover along ridge-valley catenas, from relatively dry ridge tops to flatter, moister valleys (Whittaker [1956]; Harms et al. [2001]; Bunyavejchewin et al. [2003]; Valencia et al. [2004]; Davies et al. [2005]; Wiegand et al. [2007]; Punchi-Manage et al. [2014]). Differential species occurrence along a catena is presumably due to physiological or morphological variation among species that affects responses to soil conditions (Walters and Reich [1996]; Baltzer et al. [2005]; Baraloto et al. [2007]; Engelbrecht et al. [2007]; Comita and Engelbrecht [2009]; Russo et al. [2010]). These trait differences must in-turn cause species variation in demographic performance across habitats. Specialists on a habitat should have higher fecundity, growth, or survival compared to non-specialists on that same habitat (Chesson [1985]; Givnish [1988]; Latham [1992]). Moreover, specialists on one habitat would be expected to perform best there relative to other habitats; generalists, on the other hand, are expected to perform similarly across all habitats.
We tested these demographic hypotheses of habitat association using tree census data from a fully mapped, long-term forest census plot in a species-rich tropical forest in southwestern Cameroon (Chuyong et al. [2004a]). The site is topographically variable, and many tree species have conspicuous associations with the ridge, slope, or flat valley. Indeed, 63% of tree species specialize on particular topographic subsets of the terrain (Chuyong et al. [2011]). The two predictions about variation in demography relative to topography are: 1) specialists on their favored habitat outperform other species on the same habitat; we call this the resident vs. foreign hypothesis, where resident refers to the local specialists and foreign refers to specialists of other habitats; 2) specialists perform better on their favored habitat than they do elsewhere: the home vs. away hypothesis. To test these hypotheses, we estimated growth and mortality rates of 272 species in the 50-ha forest plot and examined how rates varied across five topographic habitats along the ridge-valley catena. There were 171 species specializing on a topographic habitat, and 101 generalists, which were similarly abundant across all habitats, as detailed in Chuyong et al. ([2011]).
Methods
Study site
Korup National Park contains seasonally wet forest characteristic of southwestern Cameroon, part of the Lower Guinean forest of tropical Africa (White [1983]). The area is a former Pleistocene refugium, and tree species richness and endemism are high (Maley [1987]). Mean annual rainfall exceeds 5000 mm, with a dry season from December to February, when average monthly rainfall is <100 mm, followed by an intense wet season (Newbery et al. [1998]; Chuyong et al. [2004b]). Soils are skeletal and sandy at the surface, highly leached, and poor in nutrients (Newbery et al. [1998]; Chuyong et al. [2002]).
In the southern part of the Park, a 50-ha forest dynamics plot of 1000 m × 500 m was established at 5°03.86′ N, 8°51.17′ E (NW corner) following standardized methodology of the Center for Tropical Forest Science (Condit [1998b]). Elevation within the plot ranges from 150 to 240 m above sea level, covering diverse topography. The southern half is flat, with a valley bottom that contains a permanent stream flowing westward, whereas the northern section is steep, with gullies and large boulders (Thomas et al. [2003]; Kenfack et al. [2007]). The vegetation of the plot is a mature, closed-canopy, moist evergreen forest, with no sign of recent or ancient human disturbance. Gaps make up only 0.1% of the plot and result mostly from wind-throw (Egbe et al. [2012]). From 1996–1999, all trees with stem diameter at breast height (dbh; 1.3 m above ground) greater or equal to 1 cm were tagged, mapped, and measured at breast height, and a full re-measurement was completed in 2008–2010. The plot had 328,503 individuals in the first census, including 489 distinct taxa. Of these, 395 taxa are now fully identified species, matched with keys and herbarium specimens, including several that are newly described (Kenfack et al. [2007]), 73 taxa are identified to genus, and 21 taxa remain unknown, yet are consistently recognizable. Fewer than 500 trees remain unidentified, not sorted into any of those 489 taxa.
Based on slope, elevation, and convexity, Chuyong et al. ([2011]) identified five habitat types in the Korup plot: low-elevation depressions, low-elevation flats, high-elevation gullies, slopes, and ridge tops. We refer to these hereafter as depression, flat, gully, slope, and ridge (Figure 1). Habitat divisions were established a priori using thresholds of 165 m, 15°, and zero for mean elevation, slope, and convexity respectively. The depression habitat (mean elevation <165 m, slope <15°, and convexity <0) comprises the lowest elevation of the plot, subjected to flooding during the wet season, likely resulting in periodically anoxic soil conditions. The flat habitat (mean elevation <165 m, slope <15°, and convexity >0) is adjacent, including the low elevation portion with better drained soils. The other three habitats are at higher elevations. The slope habitat (mean elevation ≥165 m, slope ≥15°, and convexity ≥0) has the steepest inclinations, and the ridge (mean elevation ≥165 m, slope <15°, and convexity <0) includes relatively flat sections above those slopes; both have rocky, well-drained, poorly developed soil. The gully (mean elevation ≥165 m, slope ≥15°, and convexity <0) includes high elevation depressions: steep, rocky stream beds and the base of the slopes. Each 20 m × 20 m grid cell within the plot was assigned to one of the five habitats based on its topographic attributes using the methods described in Harms et al. ([2001]).
A map of topographic habitats of the Korup 50-ha plot, Cameroon, plus distribution maps for the most abundant specialist canopy tree species of each habitat. The habitats, shown in the top, left panel, are: depression (white), flat (light gray), slope (blue), gully (dark gray), ridge (green), as defined in Chuyong et al. ([2011]). Contour lines are at 2-m intervals; north is to the left. Oubanguia alata (Lecythidaceae) is a specialist of the depressions, Strombosia pustulata (Olacaceae) of flat-terrain, Uvariodendron connivens (Annonaceae) of slopes, Diospyros iturensis (Ebenaceae) of gullies, and Tabernaemontana brachyantha (Apocynaceae) of the ridge.
To identify species’ associations with topographic habitats, torus translation tests were performed on the 272 species having ≥50 stems in the plot (Chuyong et al. [2011]). For each species, the test is simply whether relative density in one habitat is higher than would be expected by random placement (where relative density is the number of trees of the species in question divided by the total number of trees of all species). The torus aspect provides a statistical test that avoids the assumption that placement of every individual is independent of other individuals; it is conservative relative to a chi-square test. Based on the torus test, 171 species had significantly higher than expected density in at least one habitat. Most were significantly associated with just one of the five habitats, but nine were positively associated with two habitats. To simplify analyses, we assigned those nine as specialists on the habitat where their density was highest, so that 171 species were designated specialists on one habitat only: 69 species associated with the depression, 31 with flat terrain, 26 with gullies, 37 with the slopes, and eight with ridge (Chuyong et al. [2011]). There were 101 species with no significant association, the generalists. Within each 20 m × 20 m grid cell, trees of a given species were considered “resident” if the grid cell was assigned to their preferred habitat, otherwise, they were considered “foreign”. The distributions of the most abundant specialists of each habitat and of all trees falling into the six association groups reveal the topographic variation (Figures 1 and 2). Sample sizes of individuals and species per habitat are given in Additional file 1. Excluded from all analyses, both in Chuyong et al. ([2011]) and here, are the 217 species with <50 individuals in the plot, comprising 3,599, or just over 1% of trees.
Distribution maps of six habitat guilds, including generalists plus specialists of the five topographic habitats. In each, trees of all species of the guild are shown together. Generalists include 101 species; depression specialists, 69 species; flat-terrain specialists, 31 species; slope specialists, 37 species; gully specialists, 26 species; and ridge specialists, 8 species. Contour lines are at 5-m intervals, and are included on each but are most visible on the map of ridge specialists.
Growth model
Estimation of diameter growth and mortality rates was based on the two censuses, and followed standard methods (Condit et al. [1999]). Growth rate for each tree (mm · y−1) was calculated as its dbh increment divided by the time interval between the two censuses, as long as both dbh were measured at the same position on the same stem. Rare outliers caused by extreme errors in dbh measurements can skew data, so we excluded all records where growth rate was >75 mm · y−1 or decreased by >4ε, where ε is dbh error estimated from double-blind measurements of a sample of trees (Condit et al. [2004]; Condit [2012]):
Therefore, we kept in the calculations many small negative growth rates which are due to routine measurement error; excluding these would bias growth rates upward.
Because individual growth rates had a highly skewed distribution, a transformation was used to normalize the data. In the past, we have used logarithmic transformation, which normalizes well but has the limitation that negative growth must be either excluded or arbitrarily set to a small positive number (Condit et al. [2006]). Because many trees show small negative growth rates caused by slight errors in dbh measurement, excluding or altering negatives causes considerable bias when growth rates are low (Condit et al. [1993]). A better alternative is to normalize with power transformation.
where g is annual dbh increment, with the addendum that
for g <0. Power transformation is a standard tool for normalizing data (e.g. Hinkley [1977]), and the option for negative numbers is a crucial advantage. The precedent for a power transformation of negative numbers is the cube root, which is defined for negatives and normalizes the gamma-distribution (Krishnamoorthy et al. [2008]). We explored powers between 0.3 and 0.5, maintaining negatives, and found that the exponent 0.45 was most effective at reducing skewness; the cube-root over-transformed, and produced skewness in the opposite direction.
Transformed growth, τ, was used in all subsequent analyses based on standard tools for regression with Gaussian responses. Results include μ τ = mean (τ) and σ τ = SD(τ) with 95% confidence limits based on the SD. We would like to present g, the annual dbh increment; however, the mean of transformed variables is not the same as the transformation of the mean:
where T is the transformation function and T−1 its inverse, and μ g is mean(g), the mean of untransformed annual dbh increment. Medians, however, do back-transform directly, so we present T−1(μ τ ) as median annual growth rate. Medians and means were quite different (as always for highly skewed data): at Korup, mean annual growth rate of all saplings (<50 mm dbh) of the 272 species we analyzed was 0.225 mm · y−1, while the median was 0.100 mm · y−1. Medians, however, are arguably a better reflection of forest growth, as for income (Spizman [2013]).
Survival model
Mortality calculations were based on annual survival probability, θ, of individual trees, calculated from the number of trees, N, in the first census, and the number of survivors, S, after t years. Survival was modeled using a binomial error distribution.
where P is the probability of the observations. The response θ was employed in statistical models (described below) in two ways: first, with a logistic transformation, logit(θ), and second, with a double-log transformation. For the latter, first define
Then ln(m) was the parameter used in modeling. Both logit and double-log methods are designed to normalize survival probabilities. We present results by back-transforming ln(m) to m, or from logit(θ) to 1 – θ; when m is low, as it is for trees, m ≅ 1 – θ is the annual mortality probability.
Statistical models for demographic hypotheses of habitat association
The two tests of habitat association are the resident-foreign hypothesis and the home-away hypothesis. The first asks whether, within a single habitat, species specializing on that habitat outperform those that do not. This could be asked one habitat at a time, but the general prediction does not distinguish between habitats and thus averages across them: do resident species in general perform better than foreign species? To specify this as a statistical model, first define X as the habitat on which a tree was located and P as the preferred habitat of its species. Next define a variable H, which is true for every individual tree growing on its preferred habitat (i.e. when X = P) and false otherwise (NULL for generalists, who are thus excluded). The foreign-resident prediction is written following the style used in the R package (R Development Core Team [2013]) with separate models for growth and mortality:
meaning that the growth or mortality of each tree is the response variable, and habitat (X) and home/away (H) are fixed-effect predictors. The term in parentheses shows that the impact of both predictors varied with species, S, thus describing the hierarchical aspect (equivalent to S being a random effect in a mixed effects model). It would be inappropriate to pool individuals, because individual-based estimates are dominated by a few abundant species and would greatly, and incorrectly, inflate statistical confidence (an error of pseudo-replication). There is no interaction term, and the single regression parameter for H reveals the advantage of resident species. Our test is whether that parameter is significantly >0 (growth) or <0 (mortality).
The home-away prediction is based on the same approach but asks how species vary across habitats: specialists should perform better on their home habitat than on other habitats. Using the definitions above, the model is written
As for Model 1, the single regression parameter for H, the home variable, is the key result. It is the mean excess performance expected for species on their own habitats, and if significantly different from zero supports the hypothesis.
Methods for estimating parameters
Mixed effects model in R
Both sets of statistical tests, resident-foreign and home-away, were executed as mixed effects models using the package lme4 in the programming language R (Bates et al. [2013]; R Development Core Team [2013]). The key feature of lme4 is multi-level modeling, allowing us to invoke species as a random effect (Gelman and Hill [2007]). When invoked for growth, lme4 calculations assume normality, and the growth model was run with τ, the transformation of growth rate. The survival model in lme4 is based the logistic transformation of θ. The output of the mixed models includes the home parameter for each, along with its standard error, providing tests of the demographic hypotheses.
Bayesian hierarchical modeling
Our main interest is demography of species, and the above mixed models produce a fixed effect estimate that is the average across all species, while the random effects are the estimates for each species. We ran Bayesian hierarchical models to estimate the mean demographic rates of individual species, as well as mean rates across species. As with the mixed models fit with lme4, the Bayesian models were run on transformed data, so the means and standard deviations are on the transformed scale. There are two levels in each model: growth (or survival) of individuals within species, and an overarching level of species within preference groups. The models were run independently for the five habitats, using transformed growth, τ (Eqs. 1 and 2), or the double-log of survival, lm(m) (Eq. 4). In each habitat, there is one parameter for mean τ of each species (or m), plus a hyper-mean and hyper-standard-deviation describing the overarching distribution across species (one pair for growth, another for survival, separately for each habitat and preference group). For growth, there must also be a within-species standard-deviation, called the residual, which we assumed to be constant for all species (the same assumption is used is mixed models in lme4); there is no residual in a survival model because the binomial distribution defines the variation. In each habitat, there were 285 growth parameters: 272 parameters for means of τ per species, 12 hyperparameters, a pair for each preference group, plus the residual parameter. For mortality, there were 272 parameters for the mean of ln(m) per species, plus the 12 hyperparameters. In addition, combined models were run: first, all habitats combined, with preference groups separated, producing a single mean for each preference group across all habitats; and second, preference groups combined but habitats separated. The means of transformed rates were back-transformed to the original scale for presentation, and thus must be interpreted as medians.
Parameters were estimated using a Markov-Chain Monte Carlo procedure based on Metropolis updates at each step (Metropolis et al. [1953]; Gelman et al. [1995]). The updates required likelihood functions giving the marginal probability of observing a single parameter value given all other parameters plus the data; for transformed growth, the likelihood functions were Gaussian for both individual species parameters and the hyperparameters, so the model consists of Gaussian species distributions nested within a Gaussian hyperdistribution. For survival, the species-level likelihoods were binomial, and the hyperdistribution was a log-normal distribution of m. Condit et al. ([2006]) used Bayesian hierarchical growth and survival models and provides further details. MCMC chains were run for 5,000 steps, with the first 1,000 discarded as burn-in. The mean of each chain provided the best estimate for a parameter, with 95% credible intervals defined by quantiles of the post-burn-in chain.
Diameter categories
The principle results are based on models in which all individuals of the 272 abundant species were included. To account for the effects of diameter on growth and mortality rate, we repeated the mixed models for saplings (all trees <50 mm dbh) and canopy trees (all >200 mm).
Results
Demography of habitat specialists and generalists across habitat types
There was no consistent performance advantage of home species, that is, no indication that species performed best on their home versus non-home habitats. This is revealed by comparing the diagonal of Table 1 against other entries on the same row. Consider, for example, the first row, which shows median demography of those species whose affinity was with the depression habitat. This group had its highest growth on the slope habitat (0.66 mm · y−1), not the depression (0.41 mm · y−1); likewise, its lowest mortality was on the ridge (1.51% · y−1) not the depression (1.73% · y−1). Indeed, none of the five preference groups had optimal growth on its home habitat, and only one group, specialists of flat terrain, had optimal survival at home.
Reading down columns corresponds to the resident-foreign test. For example, column 1 (Table 1A, 1B) shows the performance of each of the six preference groups on the depression habitat. The growth rate of the local specialists (0.41 mm · y−1) was slower than three other groups, and their mortality (1.73% · y−1) was relatively high.
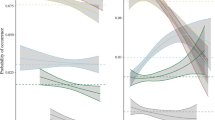
Instead, differences across habitats tended to be consistent across preference groups, as revealed by a tendency for parallel curves in Figure 3. Almost all species groups achieved their highest growth rate on the slope, and most had slow growth on depression and flats (Figure 3). Moreover, specialists groups tended to be consistent; for example, species associated with flat terrain had median growth <0.40 mm · y−1 no matter where they appeared, while species with a gully preference had growth >0.65 mm · y−1 everywhere.
Individual species demography across habitat types
Species’ growth rates were highly correlated across habitats, meaning that those with high growth on one habitat tended to have high growth on other habitats (Figure 4). Blue circles in the figures represent species specializing on the habitat of the x-axis, and red triangles specialists of the y-axis habitat. If species grew better on their favored habitat, blue points would cluster below the 1:1 line and red points above, yet this was clearly not so in any of the examples in Figure 4. For mortality rates, the expectation would be the opposite, blue above and red below, but again, this was never so.
Scatterplots showing covariation of species demographic rates across habitats. Each point represents one species, giving its growth (or mortality) rate on two habitats. Values in this figure are transformations: annual dbh increment raised to the 0.45 power and the natural log of annual mortality probability. Red triangles are specialists of the habitat of the vertical axis, and blue circles specialists of the habitat of the horizontal axis. Black circles are generalists, and small points are specialists on other habitats. A) Growth rate (transformed) on the depressions (vertical axis) vs. flat terrain. B) Growth rate (transformed) on the slopes vs. flat terrain. C) Log mortality rate on depressions (vertical axis) vs. flat terrain. D) Log mortality rate on slopes vs. flat terrain.
Abundant canopy species of each habitat preference group reinforce the same result (Figure 5). The dominant tree of the forest, Oubanguia alata, a specialist of depressions, was concentrated there, with moderate density on the low flats (Figure 2). Growth of O. alata, however, ran exactly counter to expectation: it was lowest on its home habitat (Figure 5). Indeed, none of the five abundant species had maximum growth and none had minimum mortality on its home habitat (Figure 5). Not all differences were significant, but O. alata, Strombosia pustulata, and Tabernaemontana brachyantha had significantly lower growth on their home habitats compared to at least two other habitats, and T. brachyantha had significantly higher mortality on its home habitat compared to two other habitats. Moreover, the five abundant species illustrate the tendency for consistent variation across habitat, regardless of the species’ habitat associations: all five had slow growth at low elevations, (depression and flat habitats) relative to the three ridge-slope habitats (Figure 5).
Demographic rates of five abundant species (those in Figure1), one from each habitat specialist category, across five habitats. On the left, (panels A through E) are median growth rates (mm · y−1) and on the right (panels F through J) are mortality (% · y−1). Panels A and F: depression specialist Oubanguia alata; panels B and G: flat-terrain specialist Strombosia pustulata; panels C and H: gully specialist Uvariodendron connivens; panels D and I: slope specialist Diospyros ituriensis; panels E and J: ridge specialist Tabernaemontana brachyantha. The bar is darker gray for the habitat in which each species specializes. Statistical significance based on 95% credible intervals is indicated by letters a and b at base of bars, for within-species comparisons only: habitats with a alone had a rate significantly different from habitats with b alone, but ab means no difference from either.
Test of the resident-foreign hypothesis
Precisely counter to prediction, resident species were outgrown by foreign species (Table 2). The difference was negligible and well within credible intervals, meaning there is no statistically significant variation. Mortality, on the other hand, favored resident species as predicted, but again, the variation was not statistically significant. Running the same tests separately for saplings <50 mm dbh or trees ≥200 mm yielded the same results: resident species had lower growth but higher survival, and in no case were differences significant (Table 2).
Test of the home-away hypothesis
Growth rates were again opposite the prediction, with median growth faster on away habitats compared to home habitats according to the mixed model (Table 3). The difference was negligible and not statistically distinct from zero. Mortality differed according to prediction, being lower on home habitats, but the difference was small and non-significant. Results were parallel if we included only saplings <50 mm dbh. For trees ≥200 mm dbh, species performed better when away from their home habitat based on both growth and mortality (Table 3).
Discussion
Predictions from the demographic theory of habitat association were not upheld, so differential growth and mortality of habitat specialists cannot explain how their associations with topographic habitats arose. The possibility remains that the failure to support the predictions was a problem of statistical power, but nothing about the results suggests this. Indeed, growth rates were opposite the predictions, with species on average growing more slowly on their preferred habitats. Survival did vary as predicted, favoring species on home habitats, but by tiny and non-significant amounts. Nonetheless, the habitat variation in species abundances is ecologically important, at least judging by dominant species that are concentrated in certain habitats and nearly absent on others. For example, Oubanguia alata is the dominant canopy tree across the plot, yet sparse on the slopes and ridge, but individuals on the slope and ridge in fact performed better than those in its home habitat (depression). Diospyros iturensis was five times more abundant on slopes but had higher mortality there.
Instead, demographic rates varied across habitats in a consistent way for species in each habitat association group. Growth and mortality were higher on the ridge-slope habitats, and this held across species groups and individual species. There were no indications of cross-overs in rank performance between habitats, as might be expected with habitat specialization. Other studies of tree performance have been carried out along light gradients, both experimental and observational, and similarly failed to detect cross-overs in performance ranks (Kitajima [1994]; Veneklaas and Poorter [1998]; Poorter [1999]; Kitajima and Bolker [2003]; Dalling et al. [2004]; Rüger et al. [2011a]). In another study along a soil texture gradient in a large-scale forest plot in Malaysia, many tree species distributions depend on soil type, yet specialists did not have faster growth, nor higher survival, on their preferred soils (Russo et al. [2005]).
We are forced to reject the hypothesis that demographic performance of saplings and trees in the Korup plot accounts for habitat-specific species distributions and thus must seek alternative explanations for the patterns. We suggest two classes of alternatives. First, demographic performance does matter, but we missed it in one five-year study limited to trees above 1 cm diameter. Second, negative density-dependence in demographic rates lowers demographic performance on favored sites once a species’ density is high there.
All trees in our census were at least 1 cm in diameter, well after the seedling stage. In the 50-ha plot at Barro Colorado Island, Panama, 1-cm saplings were estimated to be >10 years old (Hubbell [1998]), so habitat filtering prior to recruitment into the 50-ha census is plausible. If so, tree distribution patterns are set prior to 1-cm diameter, while larger trees show no demographic benefit in their favored habitat. A second alternative mechanism that we would miss in one five-year study is habitat filtering during unusual climatic events, such as droughts, when slope and ridge soils become exceptionally dry. Drought intensity certainly fluctuates from year-to-year and unusual droughts can have large impacts in many tropical forests (Condit [1998a]; Potts [2003]). Experimental work at other sites, including Ghana in Africa, demonstrates that species distributions are due to demographic differences in performance under drought (Veenendaal and Swaine [1998]; Engelbrecht et al. [2007]; Baltzer et al. [2008]; Comita and Engelbrecht [2009]).
A different sort of alternative hypothesis is negative density-dependence that is particularly acute on home habitats. Negative effects of high conspecific density are widely observed in tropical and temperate forests (Janzen [1970]; Connell [1971]; Condit et al. [1994]; Peters [2003]; Comita et al. [2010]; Bagchi et al. [2011]) and are likely due to pests and pathogens (Liu et al. [2012]). According to this scenario, a species is physiologically better adapted to one habitat (its home), and outperforms competitors on that habitat at low density. Better performance promotes faster population growth, and as density of the specialist builds on its home habitat relative to competitors, negative effects of enemies begin to curtail performance of the specialist. Eventually, an equilibrium results with higher density of the specialist but equal demographic performance of all species on that habitat, similar to an ideal-free distribution in animals (Fretwell and Lucas [1972]). This is distinct from the source-sink hypothesis, according to which specialists always outperform competitors from other habitats (Shmida and Wilson [1985]; Pulliam [1988]), with continual dispersal across habitats maintaining low density populations away from home.
The ubiquity of negative density-dependence suggests to us that the ideal-free distribution is a likely cause of our observations as well as the many others where demography does not differ across habitats as expected (Kitajima [1994]; Veneklaas and Poorter [1998]; Poorter [1999]; Kitajima and Bolker [2003]; Dalling et al. [2004]; Russo et al. [2005]; Yamada et al. [2007]; Rüger et al. [2011a]). But further observations of seedlings, and of all sizes in unusually dry years, are needed before we can exclude the possibility that superior performance on home habitats is common but was missed in our census. Moreover, a complete understanding of habitat specialization and niche-partitioning among tree species will require analyses of all important resources: light, moisture, and soil nutrients. These resources may covary with topography (Coomes and Grubb [2000]; Russo et al. [2012]), and it is likely that resource gradients are more complicated than a one-dimensional partitioning along topographic catenas. Our research elsewhere encompasses both light and nutrient variation (Rüger et al. [2011a], [b]; Rüger and Condit [2012]; Condit et al. [2013]), but the sharp topographic gradients at the Korup plot in Cameroon still await such evaluation.
Conclusions
Growth and mortality estimates from a five-year census in Korup reject the hypothesis that tree distributions along a ridge-valley catena are caused by demographic variation of saplings and trees. Specialists on local topographic habitats did not have improved demographic performance on their home habitats. Failure to detect demographic cross-overs has appeared in many other studies of trees, and we suggest that negative density-dependence reduces growth and survival where species reach higher densities, thus masking the superior performance of species on their home habitats.
Additional file
References
Bagchi R, Henrys P, Brown P, Bruslem FRPD, Diggle P, Gunatilleke IN, Kassim AR, Law R, Noor S, Valencia R: Spatial patterns reveal negative density dependence and habitat associations in tropical trees. Ecology 2011, 92: 1723–1729. 10.1890/11-0335.1
Baltzer JL, Thomas SC, Nilus R, Burslem DFRP: Edaphic specialization in tropical trees: Physiological correlates and responses to reciprocal transplantation. Ecology 2005, 86: 3063–3077. 10.1890/04-0598
Baltzer JL, Davies SJ, Bunyavejchewin S, Noor NSM: The role of desiccation tolerance in determining tree species distributions along the Malay-Thai peninsula. Funct Ecol 2008, 22: 221–231. 10.1111/j.1365-2435.2007.01374.x
Baraloto C, Morneau F, Bonal D, Blanc L, Ferry B: Seasonal water stress tolerance and habitat associations within four neotropical tree genera. Ecology 2007, 88: 478–489. 10.1890/0012-9658(2007)88[478:SWSTAH]2.0.CO;2
Bates D, Maechler M, Bolker B (2013) lme4: Linear mixed-effects models using S4 classes, R package version 0.999999-2. , [http://CRAN.R-project.org/package=lme4]
Bunyavejchewin S, LaFrankie JV, Baker PJ, Kanzaki M, Ashton PS, Yamakura T: Spatial distribution patterns of the dominant canopy dipterocarp species in a seasonal dry evergreen forest in western Thailand. Forest Ecol Manag 2003, 175: 87–101. 10.1016/S0378-1127(02)00126-3
Chesson PL: Coexistence of competitors in spatially and temporally varying environments: A look at the combined effects of different sorts of variability. Theor Popul Biol 1985, 28: 263–287. 10.1016/0040-5809(85)90030-9
Chuyong GB, Condit R, Kenfack D, Losos E, Sainge M, Songwe NC, Thomas DW: Korup Forest Dynamics Plot, Cameroon. In Forest Diversity and Dynamism: Findings from a Large-Scale Plot Network. Edited by: Losos EC, Leigh EG. University of Chicago Press, Chicago; 2004.
Chuyong GB, Kenfack D, Harms KE, Thomas DW, Condit R, Comita LS: Habitat specificity and diversity of tree species in an African wet tropical forest. Plant Ecol 2011, 212: 1363–1374. 10.1007/s11258-011-9912-4
Chuyong GB, Newbery DM, Songwe NC: Litter breakdown and mineralization in a central African rain forest dominated by ectomycorrhizal trees. Biogeochemistry 2002, 61: 73–94. 10.1023/A:1020276430119
Chuyong GB, Newbery DM, Songwe NC: Rainfall input, throughfall and stemflow of nutrients in a central African rain forest dominated by ectomycorrhizal trees. Biogeochemistry 2004, 67: 73–91. 10.1023/B:BIOG.0000015316.90198.cf
Comita LS, Engelbrecht BMJ: Seasonal and spatial variation in water availability drive habitat associations in a tropical forest. Ecology 2009, 90: 2755–2765. 10.1890/08-1482.1
Comita LS, Muller-Landau HC, Aguilar S, Hubbell SP: Asymmetric density dependence shapes species abundances in a tropical tree community. Science 2010, 329: 330–332. 10.1126/science.1190772
Condit R: Ecological implications of changes in drought patterns: shifts in forest composition in Panama. Clim Change 1998, 39: 413–427. 10.1023/A:1005395806800
Condit R: Tropical Forest Census Plots: Methods and Results from Barro Colorado Island, Panama and a Comparison with Other Plots. Springer, Berlin; 1998.
Condit R (2012) CTFS R Package. Smithsonian Tropical Research Institute. Accessed 21 April 2014, [http://ctfs.arnarb.harvard.edu/Public/CTFSRPackage]
Condit R, Aguilar S, Hernandez A, Pérez R, Lao S, Angehr G, Hubbell S, Foster R: Tropical forest dynamics across a rainfall gradient and the impact of an El Niño dry season. J Trop Ecol 2004, 20: 51–72. 10.1017/S0266467403001081
Condit R, Ashton PS, Manokaran N, LaFrankie JV, Hubbell SP, Foster RB: Dynamics of the forest communities at Pasoh and Barro Colorado: comparing two 50-ha plots. Philos T R Soc B 1999, 354: 1739–1748. 10.1098/rstb.1999.0517
Condit R, Ashton P, Bunyavejchewin S, Dattaraja HS, Davies S, Esufali S, Ewango C, Foster R, Gunatilleke IAUN, Gunatilleke CVS, Hall P, Harms KE, Hart T, Hernandez C, Hubbell S, Itoh A, Kiratiprayoon S, Lafrankie J, Lao S, Makana J-R, Noor SMN, Kassim AR, Russo S, Sukumar R, Samper C, Suresh HS, Tan S, Thomas S, Valencia R, Vallejo M, Villa G, Zillio T: The importance of demographic niches to tree diversity. Science 2006, 313: 98–101. 10.1126/science.1124712
Condit R, Engelbrecht BM, Pino D, Pérez R, Turner BL: Species distributions in response to individual soil nutrients and seasonal drought across a community of tropical trees. Proc Natl Acad Sci U S A 2013, 110: 5064–5068. 10.1073/pnas.1218042110
Condit R, Hubbell SP, Foster RB: Identifying fast-growing native trees from the neotropics using data from a large, permanent census plot. Forest Ecol Manag 1993, 62: 123–143. 10.1016/0378-1127(93)90046-P
Condit R, Hubbell SP, Foster RB: Density dependence in two understory tree species in a neotropical forest. Ecology 1994, 75: 671–680. 10.2307/1941725
Condit R, Pitman N, Leigh EG, Chave J, Terborgh J, Foster RB, Nunez VP, Aguilar S, Valencia R, Villa G, Muller-Landau HC, Losos E, Hubbell SP: Beta-diversity in tropical forest trees. Science 2002, 295: 666–669. 10.1126/science.1066854
Connell JH: On the Role of Natural Enemies in Preventing Competitive Exclusion in some Marine Animals and in Rain Forest Trees. In Proceedings of the Advanced Study Institute on Dynamics of Numbers in Populations. Edited by: den Boer PJ, Gradwell GR. Center for Agricultural Publishing and Documentation, Wageningen, Osterbeek, The Netherlands; 1971.
Coomes DA, Grubb PJ: Impacts of root competition in forests and woodlands: A theoretical framework and review of experiments. Ecol Monogr 2000, 70: 171–207. 10.1890/0012-9615(2000)070[0171:IORCIF]2.0.CO;2
Dalling JW, Winter K, Hubbell SP: Variation in growth responses of neotropical pioneers to simulated forest gaps. Funct Ecol 2004, 18: 725–736. 10.1111/j.0269-8463.2004.00868.x
Davies SJ, Tan S, LaFrankie JV, Potts MD: Soil-Related Floristic Variation in the Hyperdiverse Dipterocarp Forest in Lambir Hills, Sarawak. In Pollination Ecology and Rain Forest Diversity. Edited by: Roubik DW, Sakai S, Hamid A. Sarawak Studies Springer-Verlag, New York, New York; 2005.
Egbe EA, Chuyong GB, Fonge BA, Namuene KS: Forest disturbance and natural regeneration in an African rainforest at Korup National Park, Cameroon. Int J Biodivers Conserv 2012, 11: 377–384.
Engelbrecht BMJ, Comita LS, Condit R, Kursar TA, Tyree MT, Turner BL, Hubbell SP: Drought sensitivity shapes species distribution patterns in tropical forests. Nature 2007, 447: 80–83. 10.1038/nature05747
Fretwell SD, Lucas HL Jr: On territorial behavior and other factors influencing habitat distribution in birds. I. Theoretical development. Acta Biotheor 1972, 19: 16–36. 10.1007/BF01601953
Gelman A, Hill J: Data Analysis Using Regression and Multilevel-Hierarchical Models. Cambridge University Press, UK; 2007.
Gelman A, Carlin JB, Stern HS, Rubin DB: Bayesian Data Analysis. Chapman and Hall/CRC, Boca Raton; 1995.
Givnish T: Adaptation to sun and shade: A whole-plant perspective. Aust J Plant Physiol 1988, 15: 63–92. 10.1071/PP9880063
Harms KE, Condit R, Hubbell SP, Foster RB: Habitat associations of trees and shrubs in a 50-ha neotropical forest plot. J Ecol 2001, 89: 947–959. 10.1111/j.1365-2745.2001.00615.x
Hinkley D: On quick choice of power transformation. Appl Stat 1977, 26: 67–69. 10.2307/2346869
Hubbell SP: The Maintenance of Diversity in a Neotropical Tree Community: Conceptual Issues, Current Evidence, and Challenges Ahead. In Forest Biodiversity Research, Monitoring and Modeling: Conceptual Background and Old World Case Studies. Edited by: Dallmeier F, Comiskey J. Parthenon, Paris; 1998.
Janzen DH: Herbivores and the number of tree species in tropical forests. Am Nat 1970, 104: 501–528. 10.1086/282687
Kenfack D, Thomas DW, Chuyong G, Condit R: Rarity and abundance in a diverse African forest. Biodivers Conserv 2007, 16: 2045–2074. 10.1007/s10531-006-9065-2
Kitajima K: Relative importance of photosynthetic traits and allocation patterns as correlates of seedling shade tolerance of 13 tropical trees. Oecologia 1994, 98: 419–428. 10.1007/BF00324232
Kitajima K, Bolker BM: Testing performance rank reversals among coexisting species: crossover point irradiance analysis by Sack and Grubb (2001) and alternatives. Funct Ecol 2003, 17: 276–281. 10.1046/j.1365-2435.2003.07101.x
Krishnamoorthy K, Mathew T, Mukherjee S: Normal-based methods for a gamma distribution. Technometric 2008, 50: 69–78. 10.1198/004017007000000353
Latham RE: Co-occurring tree species change rank in seedling performance with resources varied experimentally. Ecology 1992, 73: 2129–2144. 10.2307/1941461
Liu X, Liang M, Etienne RS, Wang Y, Staehelin C, Yu S: Experimental evidence for a phylogenetic Janzen–Connell effect in a subtropical forest. Ecol Lett 2012, 15: 111–118. 10.1111/j.1461-0248.2011.01715.x
Maley J: Fragmentation de la forêt dense humide ouest-africaine et extension des biotopes montagnards au quaternaire récent: nouvelles données polliniques et chronologiques: implications paleoclimatiques et biogeographiques. Paleoecol A 1987, 18: 307–334.
Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E: Equation of state calculations by fast computing machines. J Chem Phys 1953, 21: 1087–1092. 10.1063/1.1699114
Newbery DM, Songwe NC, Chuyong GB: Phenology and Dynamics of an African Rainforest at Korup, Cameroon. In Dynamics of Tropical Communities. Edited by: Newbery DM, Prins HHT, Brown ND. Blackwell Science, Oxford; 1998:177–224.
Paoli GD, Curran LM, Zak DR: Soil nutrients and beta diversity in the Bornean Dipterocarpaceae: evidence for niche partitioning by tropical rain forest trees. J Ecol 2006, 94: 157–170. 10.1111/j.1365-2745.2005.01077.x
Peters HA: Neighbour-regulated mortality: the influence of positive and negative density dependence on tree populations in species-rich tropical forests. Ecol Lett 2003, 6: 757–765. 10.1046/j.1461-0248.2003.00492.x
Poorter L: Growth responses of 15 rain-forest tree species to a light gradient: the relative importance of morphological and physiological traits. Funct Ecol 1999, 13: 396–410. 10.1046/j.1365-2435.1999.00332.x
Potts MD: Drought in a Bornean everwet rain forest. J Ecol 2003, 91: 467–474. 10.1046/j.1365-2745.2003.00779.x
Pulliam HR: Sources, sinks, and population regulation. Am Nat 1988, 132: 652–661. 10.1086/284880
Punchi-Manage R, Wiegand T, Wiegand K, Getzin S, Gunatilleke CVS, Gunatilleke IAUN: Effect of spatial processes and topography on structuring species assemblages in a Sri Lankan dipterocarp forest. Ecology 2014, 95: 376–386. 10.1890/12-2102.1
R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria; 2013.
Rüger N, Berger U, Hubbell SP, Vieilledent G, Condit R: Growth strategies of tropical tree species: disentangling light and size Effects. PLoS One 2011, 6: e25330. 10.1371/journal.pone.0025330
Rüger N, Condit R: Testing metabolic theory with models of tree growth that include light competition. Funct Ecol 2012, 26: 759–765. 10.1111/j.1365-2435.2012.01981.x
Rüger N, Huth A, Hubbell SP, Condit R: Determinants of mortality across a tropical lowland rainforest community. Oikos 2011, 120: 1047–1056. 10.1111/j.1600-0706.2010.19021.x
Russo SE, Cannon WL, Elowsky C, Tan S, Davies SJ: Variation in leaf stomatal traits of 28 tree species in relation to gas exchange along an edaphic gradient in a Bornean rain forest. Am J Bot 2010, 97: 1109–1120. 10.3732/ajb.0900344
Russo SE, Davies SJ, King DA, Tan S: Soil-related performance variation and distributions of tree species in a Bornean rain forest. J Ecol 2005, 93: 879–889. 10.1111/j.1365-2745.2005.01030.x
Russo SE, Zhang L, Tan S: Covariation between understorey light environments and soil resources in Bornean mixed dipterocarp rain forest. J Trop Ecol 2012, 28: 33–44. 10.1017/S0266467411000538
Shmida A, Wilson MV: Biological determinants of spescies diversity. J Biogeogr 1985, 12: 1–20. 10.2307/2845026
Spizman LM: Developing statistical based earnings estimates: median versus mean earnings. J Legal Econ 2013, 19: 77–82.
Thomas DW, Kenfack D, Chuyong GB, Sainge NM, Losos EC, Condit RS, Songwe NC: Tree Species of Southwestern Cameroon: Tee Distributionmaps, Diameter Tables and Species Documentation of the 50-ha Korup Forest Dynamics Plot. Center for Tropical Forest Science, Washington; 2003.
Valencia R, Foster RB, Villa G, Condit R, Svenning J-C, Hernandez C, Romoleroux K, Losos E, Magard E, Balslev H: Tree species distributions and local habitat variation in the Amazon: large forest plot in eastern Ecuador. J Ecol 2004, 92: 214–229. 10.1111/j.0022-0477.2004.00876.x
Veenendaal EM, Swaine MD: Limits to Tree Species Distribution in Lowland Tropical Rainforests. In Dynamics of Tropical Communities. Edited by: Newbery DM, Prins HHT, Brown M. Blackwell Publishing, Oxford; 1998.
Veneklaas EJ, Poorter L: Growth and Carbon Partitioning of Tropical Tree Seedlings in Contrasting Light Environment. In Inherent Variation in Plant Growth: Physiological Mechanisms and Ecological Consequences. Edited by: Lambers H, Poorter H, Van Vuuren MMI. Bunkhuys Publishers, Leiden, The Netherlands; 1998.
Walters MB, Reich PB: Are shade tolerance, survival, and growth linked? Low light and nitrogen effects on hardwood seedlings. Ecology 1996, 77: 841–853. 10.2307/2265505
White F: The Vegetation of Africa. UNESCO, Paris; 1983.
Whittaker RH: Vegetation of the Great Smoky Mountains. Ecol Monogr 1956, 26: 1–80. 10.2307/1943577
Wiegand T, Gunatilleke S, Gunatilleke N: Species associations in a heterogeneous Sri Lankan dipterocarp forest. Am Nat 2007, 170: E77-E95. 10.1086/521240
Yamada T, Zuidema PA, Itoh A, Yamakura T, Ohkubo T, Kanzaki M, Tan S, Aston PS: Strong habitat preference of a tropical rain forest tree does not imply large differences in population dynamics across habitats. J Ecol 2007, 95: 332–342. 10.1111/j.1365-2745.2006.01209.x
Acknowledgments
The Korup Forest Dynamics Plot was made possible through the generous support of the National Institutes of Health award U01 TW03004 under the NIH-NSF-USDA funded International Cooperative Biodiversity Groups program, with additional financial support from the U.S. Agency for International Development’s Central Africa Regional Program for the Environment and the Smithsonian Tropical Research Institute. Financial support for the 2008 recensus was provided by the Frank Levinson Family Foundation. Analyses were supported by U.S. National Science Foundation award DEB-9806828. The Ministry of Environment and Forests, Cameroon, provided permission to conduct the field program in Korup National Park. Local administration of the project and logistic support was provided by the Bioresources Development and Conservation Programme-Cameroon, and the WWF Korup Project. We thank especially Sainge Moses for field work and Suzanne Lao for data support. The Korup Forest Dynamics Plot is part of the global network of large-scale forest demographic plots organized by Center for Tropical Forest Science and Global Forest Observatory Program of the Smithsonian Institution.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DWT conceived the 50-ha plot and raised funding. DWT and DK planned the field work, trained and supervised field workers, identified the trees, and organized data entry. RC created the final database. DK and GC conceived the main question, ran the core analysis, and drafted the manuscript. RC and SER developed and executed analyses and revised the final manuscript. All authors approved the submission.
Electronic supplementary material
40663_2014_22_MOESM1_ESM.docx
Additional file 1: Appendix 1.: Number of resident and foreign individuals on each topographic habitat in the Korup 50-ha plot, Cameroon. Resident individuals are those of species specializing on the habitat, whereas foreign individuals are those of species that specialize on one of the other habitats. Generalists belong to species with no specialization. Rare species, those with <50 individuals, were not assigned specializations due to low sample size (Chuyong et al. [2011]). The percentage of individuals in each habitat that are resident is also given. The first row gives the area out of 50 ha occupied by each habitat. Appendix 2. Number of resident and foreign species on each habitat in the Korup 50-ha plot, Cameroon. Resident species are those specializing on the habitat, whereas foreign species specialize on one of the other habitats. Generalist species are those with no specialization. Rare species, those with <50 individuals, were not assigned specializations due to low sample size (Chuyong et al. [2011]). The percentage of species in each habitat that are resident is also given. Appendix 3. Median growth (mm · y−1) and mortality (%. y−1) of Cameroonian tree species that are habitat generalists or specialists on different topographic habitats types (habitat preference), as defined in the main text. Rates with 95% confidence intervals (CI) are given for all trees (all), saplings (10–50 mm diameter at breast height; dbh), and adults (≥200 mm dbh), with the number of species (No. species) in that preference group and DBH class. All rates are the means of species medians, weighted by species, so a species with 50 individuals contributes just as much as a species with 5000 individuals. (DOCX 17 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kenfack, D., Chuyong, G.B., Condit, R. et al. Demographic variation and habitat specialization of tree species in a diverse tropical forest of Cameroon. For. Ecosyst. 1, 22 (2014). https://doi.org/10.1186/s40663-014-0022-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40663-014-0022-3