Abstract
Background
Passerida is the largest avian radiation within the order Passeriformes. Current understanding of the high-level relationships within Passerida is based on DNA–DNA hybridizations; however, the phylogenetic relationships within this assemblage have been the subject of many debates.
Methods
We analyzed the 12S ribosomal RNA gene from 49 species of Passerida, representing 14 currently recognized families, to outline the phylogenetic relationships within this group.
Results
Our results identified the monophyly of the three superfamilies in Passerida: Sylvioidea, Muscicapoidea and Passeroidea. However, current delimitation of some species is at variance with our phylogeny estimate. First, the Parus major, which had been placed as a distinct clade sister to Sylvioidea was identified as a member of the super family; second, the genus Regulus was united with the Sturnidae and nested in the Muscicapoidea clade instead of being a clade of Passerida.
Conclusion
Our results were consistent with Johansson’s study of the three superfamilies except for the allocation of two families, Paridae and Regulidae.
Similar content being viewed by others
Background
Passeriformes is the largest group of birds in the world, containing nearly 60% of all bird species found on all continents except Antarctica (Sibley and Monroe 1990). The classification of the major groups of passerine birds has long been an issue of debate among avian systematists (Voous 1985; Sibley and Ahlquist 1990; Ericson and Johansson 2003; Barker et al. 2004). Phylogenetic analyses based on morphological characteristics are particularly difficult because of the great similarity and a high level of convergent evolution among passerine families (Beecher 1953; Tordoff 1954; Ames 1971; Raikow 1978). This limits the potential to outline high-level relationships within this assemblage using morphology. Sibley and Ahlquist (1990) finished the first comprehensive molecular study based on DNA-DNA hybridization data and suggested a different phylogenetic relationship from previously morphological characteristics based on using DNA-DNA hybridization. Their results support the traditional delimitation of oscines (suborder Passeri) and sub-oscines (suborder Tyranni) and, as well, they suggested that the oscines consisted of two sister groups, the Corvida, a clade that is primarily an Australo–Papuan group of “crow-like” birds and the Passerida, primarily a Northern Hemisphere group (Sibley and Monroe 1990). Passerida is the largest group within Passeriformes, which is further divided into three “superfamilies”, the Muscicapidae, such as waxwings, dippers, thrushes, Old World flycatchers, starlings and mockingbirds; the Sylvioidea, such as nuthatches, tits, wrens, swallows, bulbuls, babblers and sylviine warblers and the Passeroidea, i.e., larks, pipits, wagtails, waxbills, weavers, finches, sparrows, cardinals, tanagers, wood warblers and blackbirds. Although the Muscicapoidae was considered to be the sister group of the other two groups (Sibley and Monroe 1990), this relationship was not corroborated by reanalysis of the DNA-DNA hybridization data set (Harshman 1994). Furthermore, monophyly of Passeroidea and Sylvioidea could not be confirmed by a more sophisticated experimental design and rigorous statistical analyses of the DNA-DNA hybridization data (Sheldon and Gill 1996). In recent years, Sibley and Alquist’s taxonomy of birds has become the framework for many ecological and phylogenetic studies (e.g. Starck and Ricklefs 1998; Bennett and Owens 2002; Hawkins et al. 2006), however, their “Tapestry” also has been subjected to many criticisms, ranging from non-reproducibility (Mindell 1992) to sparse sampling of the complete distance matrix (Lanyon 1992).
Many studies have been conducted based on mitochondrial and nuclear gene sequence data to clarify the molecular phylogenetic relationships within passerines. However, the basal relationships of Passerida remain unresolved. For example, some disputes focused in the monophyly of Muscicapoidae (Cibois and Cracraft 2004; Voelker and Spellman 2004) and Sylvioidea (Spicer and Dunipace 2004), the origin of the oscine (Barker et al. 2002; Ericson et al. 2002; Fuchs et al. 2006) and the classification status of the Regulidae (Sturmbauer et al. 1998; Spicer and Dunipace 2004), Paridae (Ericson and Johansson 2003; Treplin et al. 2008; Fregin et al. 2012) and Alaudidae (Barker et al. 2002; Alström et al. 2006; Fregin et al. 2012). Johansson et al. (2008) presented a new phylogeny of Passerida based on three nuclear introns, showing that the Passerida is divided into nine groups with unresolved relationships. In addition, the following new molecular phylogenetic relationships at lower taxonomic levels have been identified: (1) the Muscicapinae and Turdinae (as circumscribed in Sibley and Monroe 1990) are not monophyletic (Voelker and Spellman 2004) and the Saxicolinae is paraphyletic (Sangster et al. 2010), (2) a revised classification of Parulidae (Lovette et al. 2010) and (3) the family of Locustellidae is non-monophyletic (Alström et al. 2011). DNA sequence-based studies also indicated that several of the taxa that were included in the Passerida did not belong to this radiation according to the taxonomy of Sibley and Monroe (1990). For example, the Tibetan Ground-Jay (Pseudopodoces humilis), which in previous classifications was placed among the crows (e.g. Riley 1930; Hume 1871; Monroe and Sibley 1993; Cibois et al. 1999), has recently been shown to be a ground-living tit (Paridae) (James et al. 2003). The uncertainty and conflict in these studies show a clear need to increase both the number of independent loci sequenced and the extent of taxon sampling to help resolve difficult nodes.
Ribosomal genes are considered to be modestly evolving sequences (Pereira and Baker 2006) and are suitable for high-level phylogenetic research (Hedges 1994; Vun et al. 2011). In our study, we applied a mitochondrial locus, i.e., the 12S ribosomal RNA gene, as genetic marker to explore several issues relating to Passerida. Our primary objective was to examine the basal relationships within the Passerida and test the monophyly of the three superfamilies. In addition, we investigated the taxonomic position of several taxa of uncertain phylogenetic affinities.
Methods
Sampling, amplification and sequencing
We obtained frozen tissue samples from two individuals of 15 species from collections of the Life Science Museum of Hebei Normal University (See Additional file 1: Table S1). The total DNA was extracted followed the procedure described by Sambrook and Russell (2001). The 12S ribosomal RNA gene was amplified in a 50 μL amplification reaction using a PCR kit (TransGen Biotech Co. (Beijing)). The PCR reactions were amplified using the following profile: denaturation at 94°C for 1 min followed by 30 cycles consisting of a 45-s 94°C denaturation step, a 40-s 55°C annealing step, a 1-min 72°C extension step and a final extension of 5 min at 72°C. The PCR products were verified by electrophoresis and purified using a gel extraction kit according to the instructions of the manufacturer. The purified PCR amplicons were cycle-sequenced at Sangon Biotech Co. (Shanghai). The primer pair used for the DNA amplification and sequencing was designed by Sorenson et al. (1999): L1753, 5′-AAACTGGGATTAGATACCCCACTAT-3′; and H2294, 5′-TTTTCAGGCGTAAGCTGAATGCTT-3′. All sequences were deposited in GenBank (Additional file 1: Table S1). We also downloaded the 12S ribosomal gene sequence data of 34 closely related ingroup species from GenBank to analyze together with our data (Additional file 1: Table S1). In total 49 species were sampled to represent 14 ingroup families of Passerida. Redbacked Shrike (Lanius collurio) and Rook (Corvus frugilegus) were used as outgroups in this analysis.
Phylogenetic analyses
All 51 species of the 12S ribosomal RNA sequence were aligned using Clustal X 1.81 (Thompson et al. 1997). The sequence alignments were unambiguous and verified visually. For all the phylogenetic analyses, the gaps were classified as missing data. These analyses were performed on a reduced sequence dataset (435 bp) after cutting off both ragged sides. The genetic distance and the statistics of the nucleotide composition of all the taxa were computed in MEGA 4.0 (Tamura et al. 2007). The phylogenetic relationships were reconstructed using maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI). The MP and ML analyses were performed in PAUP*4.0b10a (Swofford 2002). The MP analysis was carried out using heuristic searches with 1000 random addition sequence replicates, tree bisection reconnection (TBR) branch swapping and the transitions and transversions were given equal weight. The ML analysis was based on the best-fit substitution model, which was selected using the Akaike Information Criterion in Modeltest version 3.7 (Posada and Crandall 1998). For the ML analysis, a heuristic search with the TBR branch swapping algorithm and 100 random addition replicates were used. To assess nodal reliability, bootstrap analyses were conducted with 1000 replicates for the tree topologies of MP and ML. The BI analysis was performed using MrBayes version 3.1.2 (Huelsenbeck and Ronquist 2001). The BI searches used Metropolis-coupled Markov chain Monte Carlo sampling with one cold and three heated chains running for 4000000 generations with a sampling frequency of 100. The generations sampled before the chain reached stationarity were discarded as “burnin”. Subsequently, the posterior probabilities were derived from the 50% majority rule consensus of all the trees retained.
Results
Sequence characteristics and genetic diversity
The sequences obtained from the 12S rRNA locus (partial sequence) of 15 species representing 7 families varied in length from 435 bp in the Lanceolated Warbler (Locustella lanceolata) to 562 bp in the Pallas’s Warbler (Phylloscopus proregulus) (median = 523 bp). When these sequences were aligned with previous published sequences of the 34 closely related species, there were 312 conserved sites, 198 variable sites, including 153 parsimony information sites and 45 singleton sites, among the 515 sites, including gaps, in a total of 49 ingroup sequences. The nucleotide frequencies were 0.296 (A), 0.190 (T), 0.287 (C) and 0.227 (G). The 12S ribosomal RNA sequences were enriched in adenine residues and deficient in thymine residues; however, the GC and AT contents were nearly equal (51.9 and 49.1%). The transition/transversion rate ratios were k 1 = 2.845 (purines) and k 2 = 5.076 (pyrimidines). All positions containing gaps and missing data were eliminated from the dataset (Complete-deletion option). Pairwise sequence divergence within the ingroup at this locus varied from 0.1% (between White Wagtail (Motacilla alba) and Gray Wagtail (Motacilla cinerea)) to 16.5% (between Yellowbrowed Warbler (Phylloscopus inornatus) and Grosbeak Weaver (Amblyospiza albifrons)).
Phylogenetic analysis
In the MP analysis, 12 equally parsimonious trees were obtained using a branch-swapping algorithm. The tree length was 911, the consistency index (CI) 0.3061 and the retention index (RI) 0.5808. Our bootstrap analysis showed large numbers of well-supported nodes (see Figure 1). Of the 48 nodes that were retained in the strict consensus of the equally parsimonious tree, 37 (77%) were recovered in ≥ 50%, 28 (58%) in ≥ 70% and 24 (50%) in ≥ 90% of bootstrap replicates.
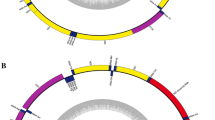
The maximum-Parsimony tree (strict consensus of 12 trees, L = 911 steps) obtained from the passerine analyses of the 12S ribosomal data. The numbers on the branches are the nodal support data. The proportion of bootstrap replicates in which a given node was recovered is indicated by branch thickness: a thickened branch indicates BP ≥ 90%.
ML analysis of the sequence evolution was calculated with the hierarchical likelihood ratio test (hLRTs) using Modeltest 3.7 and yielded the GTR + I + G model as the best-fit model. The same model was used for the BI analysis. These two analyses obtained the same topology of the phylogenetic trees but with minor differences in the value of the bootstrap support (BP) and posterior probabilities (PP), such that only the ML tree is presented (Figure 2). The majority of the branch nodes indicated high BP and PP values. Of the 49 nodes retained in the consensus ML tree, 36 (73%) were recovered in ≥ 50%, 28 (57%) in ≥ 70% and 25 (51%) in ≥ 90% of bootstrap replicates. The relative number of nodes in the BI was 37 (78%) ≥ 0.50, 30 (61%) ≥ 0.70 and 25 (51%) ≥ 0.90.
Maximum-likelihood tree (−lnL = 4869.58551) obtained from the passerine analyses of 12S ribosomal data. The numbers on the branches are the nodal support data (the proportion of bootstrap replicates/posterior probability of Bayesian inference). The proportion of bootstrap replicates in which a given node was recovered is indicated by branch thickness: a thickened branch indicates BP ≥ 90% and PP ≥ 0.90.
In MP and ML tree topologies (Figures 1 and 2), all species were divided into three major clades: Sylvioidea, Muscicapoidea and Passeroidea. Sylvioidea sistered to Passeroidea and Muscicapoidea clade sistered to both Sylvioidea and Passeroidea. The mean genetic distances calculated using the Kimula 2-Parameter model in the three major clades are as follows: Passeroidea/Sylvioidea 0.091, Passeroidea/Muscicapoidea 0.097 and Sylvioidea/Muscicapoidea 0.106.
Among the three phylogenetic trees, the topology structure of Muscicapoidea clade was congruent, whereas the topology of Passeroidea clade in the MP tree was different from the ML and BI trees. In the MP tree, the Orange-bellied Flowerpecker (Dicaeum trigonostigma) and Collared Sunbird (Anthreptes collaris) constituted one basal branch of the Passeroidea clade, whereas they were two separate clades in the ML and Bayesian trees. The location of the Amblyospiza albifrons was different. It was the sister species to Passer and Emberiza in the MP tree but clustered with the Loxia and Carduelis in the ML and BI trees. The topologies of the Sylvioidea were largely different in the three trees, although they were composed of the same species (Figures 1 and 2). In the MP tree, the Sylvioidea clade consisted of three separated clades. Phylloscopus was placed at the deepest branch as a basal lineage and sistered to the other two branches (Figure 1). The first branch included sampled species of Acrocephalus, Apalis, Cisticola, Parus, Pycnonotus, Panurus, Aegithalos and Eremophila, while the other branch contained Locustella and two sister groups. One sister group comprised Anthoscopus, Psalidoprocne and Tachycineta and the other sister group included representatives of Leiothrix and Zosterops. In the ML tree, the Sylvioidea comprised two separate branches with strong support (BP = 93%, PP = 0.96). The first branch, which diverged early, consists of two sister groups, Eremophila/Pycnonotus and Parus/Cisticola and the other branch consists of the rest of the species (Figure 2). In the latter clade, Locustella, Acrocephalus and Aegithalos comprised three external clades with two interior clades nested inside. One interior clade comprised Panurus, Anthoscopus, Psalidoprocne, Tachycineta and Phylloscopus and the other interior clade included Apalis, Leiothrix and Zosterops (Figure 2). In addition, a striking arrangement was observed for Pseudopodoces humilis, which was sampled to represent Paridae. Pseudopodoces humilis was nested in the clade corresponding to Muscicapidae and was placed as a sister taxa to species of Muscicapidae with strong support (91% in the BP analysis and 94% in both the MP and ML analyses and PP = 0.98 in the BI analysis).
Discussion and conclusions
Phylogeny of the three superfamilies
In our results, all sampled members of the Passerida were clustered together in the MP, ML and Bayesian trees. Our results identified the monophyly of the three superfamilies. Both Sylvioidea and Muscicapoidea obtained strong support in the ML and BI analyses (BP > 90% in the ML analysis and PP > 90% in the BI analysis). They were also supported by the MP analysis (BP > 80%). The Passeroidea clade was weakly supported by three analyses (BP = 56% in the MP analysis and 57% in the ML analysis and PP = 69% in the BI analysis). The classification of Johansson et al. (2008) was supported by our results with two exceptions. First, the Parus major, which had been placed as a distinct clade sister to Sylvioidea (Johansson et al. 2008; Treplin et al. 2008) was identified as a member of the super family; second, the Regulidae, Regulus, classified by Jonsson and Fjeldsa (2006) as a deep sole clade in the Passerida, was united with the Sturnidae and nested in the Muscicapoidea clade.
Barker et al. (2002) suggested that the Sylvioidea was the earliest split of the Passerida. In the topology of the phylogeny of Johansson et al. (2008) the Passeriodea was the basal clade, but we found that the Muscicapoidea was a basal lineage for the sampled taxa in this study. Our results are largely consistent with the conclusions of recent studies based on the mitochondrial genome (Nabholz et al. 2010), which suggested that the Muscicapoidea was the deepest clade sister to the clade grouping Sylvioidea and Passeroidea. However, in their phylogenetic trees, the Paridae represented a deep, isolated lineage within the Passerida.
Relationships within Muscicapoidea
The monophyly of the Muscicapoidea was revealed with strong support and the structure of this clade was congruent in the three trees. The species of Turdinae constitute the deepest branch within the superfamily with strong support and sister to all the other Muscicapoidea taxa. These results are not consistent with the monophyly of Muscicapidae, the “core muscicapoid” group that was defined by Barker et al. (2002). This was not the first time that the status of the Turdinae had been questioned. Voelker and Spellman (2004) determined that the Turdinae was not a part of the Muscicapidae but was sister to a Sturnidae and Cinclidae clade based on the nuclear c-mos gene and mitochondrial loci data. Regulidae was defined as a sole clade by Johansson et al. (2008), but in this study it was placed in the Muscicapidae clade and grouped with the Sturnus in the trees constructed from our data. The position of the Regulidae has been argued in previous studies. Sturmbauer et al. (1998) determined that the kinglets were in the Sylvioidea and were a sister to a clade comprising Parus and Phylloscopus, Spicer and Dunipace (2004) placed it in the Corvoidea, while Fregin et al. (2012) arranged it in Passeroidea. Our results show that the Regulidae should belong to the Muscicapoidea, an arrangement that agrees with the study of Beresford et al. (2005) based on sequence data of a nuclear RAG gene. In addition, the Muscicapini (Ficedula and Muscicapa) was found to be nested in the Saxicolinae, which indicates the paraphyly of these taxa (Sangster et al. 2010). The closest relationship of the genus Tarsiger and the genus Ficedula (Outlaw and Voelker 2006) was also supported by our results. Pseudopodoces humilis had ever been placed in the clade of Corvoidea (Sibley and Monroe 1990) and could be removed into the clade of Paridae (James et al. 2003). Although we have one shorter marker we corroborated that Pseudopodoces humilis does not belong to the Corviods, but is a sister to the Muscicapini instead of grouping it with Parus.
Relationships within the Passeroidea
We revealed the monophyly of the Passeroidea, although this clade receives only weak support (BP was 56% in the MP analysis and 57% in the ML analyses and the PP = 0.69 in the BI analysis). In the major clade corresponding to the “Passeroidea” defined by Johansson et al. (2008), the Nectariniidae, represented by Dicaeum trigonostigma and Anthreptes collaris, was the sister group to the other examined sparrows with one single (Figure 1, MP tree) or two separate clades (Figure 2, ML and BI trees). The other sampled taxa in this major clade were divided into two groups. All sampled taxa in these two groups have same positions in the three trees except Amblyospiza albifrons, which was clustered with Loxia and Carduelis in the MP tree but was grouped with Passer and Emberiza in the ML and Bayesian trees. We found that the Motacillidae is closely related with Fringillidae. This arrangement was also recovered in a previous study (Beresford et al. 2005). Our results also show the polyphyly of the Emberizinae, as well as the sampled taxa of this subfamily, divided into two groups of the Passeroidea clade. This discovery was not consistent with Sibley and Ahlquist (1990), but was identical to the findings of Yuri and Mindell (2002), which were based on mitochondrial sequence data.
Relationships within the Sylvioidea
Although the major clade corresponding to the “Sylvioidea” included the same taxa and obtained strong support in the three phylogenetic trees (Figures 1 and 2), the relationships within this assemblage were largely different between the MP analysis and the other analyses (the ML and BI analyses obtained similar results). For example, the basal lineage was Phylloscopus in the MP tree, whereas this location was occupied by the group consisting of Eremophila, Pycnonotus, Parus and Cisticola in the trees constructed from the ML and BI analyses (see Figures 1 and 2). The Sylvioidea clade in these trees included the sampled taxa representing the Sylviidae (Phylloscopus, Acrocephalus, Locustella, Panurus and Leiothrix), Aegithalidae (Aegithalos), Hirundinidae (Tachycineta and Psalidoprocne), Pycnonotidae (Pycnonotus), Cisticolidae (Cisticola and Apalis), Zosteropidae (Zosterops) and Paridae (Anthoscopus and Parus). This clade also included the horned lark (Eremophila alpestris), the representative of Eremophila.
We also found some similarities in these three analyses despite the high divergence in the clade Sylvioidea. First, the non-monophyly of the Paridae was identified. The Parus and Anthoscopus were sisters to the Cisticola and Hirundinidae (Psalidoprocne and Tachycineta) respectively and their relationships were highly supported. Second, the Leiothrix was sister to Zosterops and demonstrated the close relationship of the two groups. This relationship received strong support in three analyses. Third, the polyphyly of the Sylviidae (Cibois et al. 1999) was supported by our results, whereas the Locustella was located in a basal separate clade. The other sampled genera of the Sylviidae, Phylloscopus, Acrocephalus, Leiothrix and Panurus, were placed in several separate clades. Finally, the polyphyly of the Cisticolidae was confirmed for the two genera of this family, Apalis and Cisticola. They were located in different branches of this assemblage. We must conclude that the relationships within Sylvioidea are poorly resolved in our study, a situation that may be related to the rapid radiation of the families within this assemblage (Fregin et al. 2012).
References
Alström P, Ericson PG, Olsson U, Sundberg P (2006) Phylogeny and classification of the avian superfamily Sylvioidea. Mol Phylogenet Evol 38:381–397
Alström P, Fregin S, Norman JA, Ericson PGP, Christidis L, Olsson U (2011) Multilocus analysis of a taxonomically densely sampled dataset reveal extensive non-monophyly in the avian family Locustellidae. Mol Phylogenet Evol 58:513–526
Ames PL (1971) The morphology of the syrinx in passerine birds. Bull Peabody Mus Nat Hist 37:1–194
Barker FK, Barrowclough GF, Groth JG (2002) A phylogenetic hypothesis for passerine birds: taxonomic and biogeographic implications of an analysis of nuclear DNA sequence data. Proc R Soc Lond B 269:295–308
Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J (2004) Phylogeny and diversification of the largest avian radiation. Proc Nat Acad Sci USA 101:11040–11045
Beecher WJ (1953) A phylogeny of the oscines. Auk 70:270–333
Bennett PM, Owens IPF (2002) Evolutionary Ecology of Birds. Life Histories, Mating Systems and Extinction. Oxford University Press, New York
Beresford P, Barker FK, Ryan PG, Crowe TM (2005) African endemics span the tree of songbirds (Passeri): molecular systematics of several evolutionary ‘enigmas’. Proc R Soc Lond B 272:849–858
Cibois A, Cracraft J (2004) Assessing the passerine “Tapestry”: phylogenetic relationships of the Muscicapoidea inferred from nuclear DNA sequences. Mol Phylogenet Evol 32:264–273
Cibois A, Pasquet E, Schulenberg TS (1999) Molecular systematics of the Malagasy babblers (Passeriformes: Timaliidae) and warblers (Passeriformes: Sylviidae), based on cytochrome b and 16S rRNA sequences. Mol Phylogenet Evol 13:581–595
Ericson PGP, Johansson US (2003) Phylogeny of Passerida (Aves: Passeriformes) based on nuclear and mitochondrial sequence data. Mol Phylogenet Evol 29:126–138
Ericson PGP, Christidis L, Cooper A, Irestedt M, Jackson J, Johansson US, Norman JA (2002) A Gondwanan origin of passerine birds supported by DNA sequences of the endemic New Zealand wrens. Proc R Soc Lond B 269:235–241
Fregin S, Haase M, Olsson U, Alström P (2012) New insights into family relationships within the avian superfamily Sylvioidea (Passeriformes) based on seven molecular markers. Evol Biol 12:157
Fuchs J, Fjeldsa J, Bowie RC, Voelker G, Pasquet E (2006) The African warbler genus Hyliota as a lost lineage in the Oscine songbird tree: molecular support for an African origin of the Passerida. Mol Phylogenet Evol 39:186–197
Harshman J (1994) Reweaving the tapestry: what can we learn from Sibley and Ahlquist (1990)? Auk 111:377–388
Hawkins BA, Diniz-Filho JAF, Jaramillo CA, Soeller SA (2006) Post-Eocene climate change, niche conservatism, and the latitudinal diversity gradient of New World birds. J Biogeogr 33:770–780
Hedges SB (1994) Molecular evidence for the origin of birds. Proc Natl Acad Sci USA 91:2621–2624
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755
Hume A (1871) Stray notes on ornithology in India. Ibis 3:403–413
James HF, Ericson PGP, Slikas B, Lei F-M, Gill FB, Olson SL (2003) Pseudopodoces humilis, a misclassified terrestrial tit (Aves: Paridae) of the Tibetan Plateau: evolutionary consequences of shifting adaptive zones. Ibis 145:185–202
Johansson US, Fjeldsa J, Bowie RCK (2008) Phylogenetic relationships within Passerida (Aves: Passeriformes): a review and a new molecular phylogeny based on three nuclear intron markers. Mol Phylogenet Evol 48:858–876
Jonsson KA, Fjeldsa J (2006) A phylogenetic supertree of oscine passerine birds (Aves: Passeri). Zool Scr 35:149–186
Lanyon SM (1992) Review: phylogeny and classification of birds: a study in molecular evolution. Condor 94:304–307
Lovette IJ, Perez-Eman JL, Sullivan JP, Banks RC, Fiorentino I, Cordoba-Cordoba S, Echeverry-Galvis M, Barker FK, Burns KJ, Klicka J, Lanyon SM, Bermingham E (2010) A comprehensive multilocus phylogeny for the wood-warblers and a revised classification of the Parulidae (Aves). Mol Phylogenet Evol 57:753–770
Mindell DP (1992) DNA–DNA hybridization and avian phylogeny. In review: phylogeny and classification of birds: a study in molecular evolution (C. G. Sibley and J. E. Ahlquist). Syst Biol 41:126–134
Monroe BL Jr, Sibley CG (1993) A World Checklist of Birds. Yale University Press, London
Nabholz B, Jarvis ED, Ellegren H (2010) Obtaining mtDNA genomes from next-generation transcriptome sequencing: a case study on the basal Passerida (Aves: Passeriformes) phylogeny. Mol Phylogenet Evol 57:466–470
Outlaw DC, Voelker G (2006) Systematics of Ficedula flycatchers (Muscicapidae): a molecular reassessment of a taxonomic enigma. Mol Phylogenet Evol 41:118–126
Pereira SL, Baker AJ (2006) A mitogenomic timescale for birds detects variable phylogenetic rates of molecular evolution and refutes the standard molecular clock. Mol Phylogenet Evol 23:1731–1740
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Raikow RJ (1978) Appendicular myology and relationships of the New World nine-primaried oscines (Aves: Passeriformes). Bull Carnegie Mus Nat Hist 7:1–43
Riley JH (1930) Birds collected in Inner Mongolia, Kansu and Chili by the National Geographic Society’s central China Expedition under the direction of F. R. Wulsin. Proc U S Nat Mus 77:20–21
Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York
Sangster G, Alström P, Forsmark E, Olsson U (2010) Multi-locus phylogenetic analysis of Old World chats and flycatchers reveals extensive paraphyly at family, subfamily and genus level (Aves: Muscicapidae). Mol Phylogenet Evol 57:380–392
Sheldon FH, Gill FB (1996) A reconsideration of songbird phylogeny, with emphasis on titmice and their sylvioid relatives. Syst Biol 45:473–495
Sibley CG, Ahlquist JE (1990) Phylogeny and Classification of Birds: A Study in Molecular Evolution. Yale University Press, London
Sibley CG, Monroe BL Jr (1990) Distribution and Taxonomy of Birds of the World. Yale University Press, London
Sorenson MD, Ast JC, Dimcheff DE, Yuri T, Mindell DP (1999) Primers for a PCR-based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol Phylogenet Evol 12:105–114
Spicer GS, Dunipace L (2004) Molecular phylogeny of songbirds (Passeriformes) inferred from mitochondrial 16S ribosomal RNA gene sequences. Mol Phylogenet Evol 30:325–335
Starck JM, Ricklefs RE (1998) Avain Growth and Development. Oxford University Press, New York
Sturmbauer C, Berger B, Dallinger R, Foger M (1998) Mitochondrial phylogeny of the genus Regulus and implications on the evolution of breeding behavior in sylvioid songbirds. Mol Phylogenet Evol 10:144–149
Swofford DL (2002) PAUP*: Phylogenetic Analysis using Parsimony and Other Methods: Beta Version. Sinauer Associates, Massachusetts
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence: alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Tordoff HB (1954) A systematic study of the avian family Fringillidae based on the structure of the skull. Misc Publ Mus Zool Univ Mich 81:1–42
Treplin S, Siegert R, Bleidorn C, Thompson HS, Fotso R, Tiedemann R (2008) Molecular phylogeny of songbirds (Aves: Passeriformes) and the relative utility of common nuclear marker loci. Cladistics 24:328–349
Voelker G, Spellman GM (2004) Nuclear and mitochondrial DNA evidence of polyphyly in the avian superfamily Muscicapoidea. Mol Phylogenet Evol 30:386–394
Voous KH (1985) Passeriformes. In: Campbell B, Lack E (eds) A Dictionary of Birds. T. and A.D. Poyser, Carlton, UK, pp 440–441
Vun VF, Mahani MC, Lakim M, Ampeng A, Md-Zain BM (2011) Phylogenetic relationships of leaf mokeys (Presbytis:Colobinae) based on cytochrome b and 12S rRNA genes. Genet Mol Res 10:368–381
Yuri T, Mindell DP (2002) Molecular phylogenetic analysis of Fringillidae, “New World nine-primaried oscines” (Aves: Passeriformes). Mol Phylogenet Evol 23:229–243
Acknowledgements
This research was supported by the National Natural Science Foundation of China (NSFC, 31000191, 31330073), the Postdoctoral Science Foundation of China (2011 M500537), the Natural Science Foundation of Hebei Province (NSFHB, 2012205018), the Natural Science Foundation of the Department of Education, Hebei Province (YQ2014024) to D. Li, and NSFHB (2013205018) to Y. Wu.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YW and DL designed the experiments and LW conducted the experiments. LW, YS, JL and YL analyzed the data and finished the earlier draft of the manuscript. All authors read and approved the final manuscript.
Additional file
Additional file 1: Table S1.
The species in which the 12S ribosomal RNA genes were sequenced for this study. Table S2. List of sample sequences retrieved from GenBank used in the study.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Wu, L., Sun, Y., Li, J. et al. A phylogeny of the Passerida (Aves: Passeriformes) based on mitochondrial 12S ribosomal RNA gene. Avian Res 6, 1 (2015). https://doi.org/10.1186/s40657-015-0010-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40657-015-0010-5