Abstract
Background
d-Galactose dehydrogenase (GalDH; EC 1.1.1.48) belongs to the family of oxidoreductases that catalyzes the reaction of β-d-galactopyranose in the presence of NAD+ to d-galacto-1,5-lactone and NADH. The enzyme has been used in diagnostic kits to neonatal screen for galactosemia diseases. This article reports the partitioning optimization of recombinant Pseudomonas fluorescens GalDH in aqueous two-phase systems (ATPS).
Methods
Preliminary two-phase experiments exhibited that the polyethylene glycol (PEG) concentration, pH value, and concentration of salt had a significant influence on the partitioning efficiency of recombinant enzyme. According to these data, response surface methodology (RSM) with a central composite rotatable design (CCRD) was performed to condition optimization.
Results
The optimal partition conditions were found using the 14.33% PEG-4000 and 11.79% ammonium sulfate with pH 7.48 at 25°C. Yield, purity, recovery, and specific activity were achieved 92.8%, 58.9, 268.75%, and 373.9 U/mg, respectively. PEG and ammonium sulfate concentration as well as pH indicated to have a significant effect on GalDH partitioning. Enzyme activity assay and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis demonstrated the suitability of predicted optimal ATPS as well. The Km and molecular weight values for the purified GalDH were 0.32 mM and 34 kDa, respectively.
Conclusions
Ultimately, our data showed the feasibility of using ATPS for partitioning and recovery of recombinant GalDH enzyme.
Similar content being viewed by others
Background
Liquid-liquid extraction using aqueous two-phase systems (ATPS) has been applied for recovery and purification of many industrial enzymes [[1],[2]]. When two aqueous solutions of certain incompatible substances, such polyethylene glycol (PEG) and dextran or PEG and salt, are mixed above a critical concentration, two-phase separation occurs. Separation techniques based on two-phase partitioning have proved to be suitable tools for recovery of biomolecules. Compared with the traditional techniques, ATPS have the advantages such as ensuring high values of the purification parameters, preserving the targeted biomolecules, yielding separation performance, and ease to scale-up. Successful applications of ATPS for downstream processing of proteins on industrial scales have been demonstrated [[3],[4]].
d-Galactose dehydrogenase (GalDH; d-galactose: NAD+ oxidoreductase; EC 1.1.1.48) belongs to the family of oxidoreductases that catalyzes the dehydrogenation reaction of β-d-galactopyranose in the presence of NAD+ to d-galacto-1,5-lactone and NADH. The kinetic mechanism of Bi-Bi has been determined for this enzyme, with the NAD+ binding first to the enzyme. The substrates of GalDH are d-galactose and NAD+, whereas its products are D-galactono-1,4-lactone, NADH, and H+ [[5]]. GalDH has been identified in plants (e.g., green peas and Arabidopsis thaliana), algae (e.g. Iridophycus flaccidum), bacteria, and mammals. However, GalDH from Pseudomonas fluorescens bacterium is the best investigated enzyme, as its recombinant form has been produced in Escherichia coli [[6],[7]]. GalDH is a significant tool for the measurement of β-d-galactose, α-d-galactose, and lactose as well. The enzyme has been used in diagnostic kits to screen blood serum of neonates for galactosemia diseases [[8]]. Galactosemia is an inborn metabolic disorder that without strict dietary control results in mental retardation, microcephaly, and seizures. Newborn screening using GalDH is a simple method which has proved sensitive, reliable, rapid, and cheap compared to other methodologies [[9]]. This enzyme has been purified by conventional methods including ammonium sulfate precipitation followed by chromatography which are usually time-consuming and expensive [[6],[8]]. Owing to the commercial importance of GalDH, developing the efficient and scalable alternative methods for downstream processing is of great interest. In this work, we aimed to use ATPS technology for partitioning of P. fluorescens GalDH. The best partition conditions are generally achieved by systematic variation of different parameters such as temperature, pH, size and concentration, and type of polymer and salt. However, despite the apparent simplicity, partition of compounds is very complex due to the several factors involved. In fact, the classical optimization approach varying the level of one parameter at a time, while holding the rest of the variables constant, is generally time-consuming [[10]]. For these reasons, mathematical modeling has been utilized to identify parameters mainly those that affect the partition of proteins in ATPS [[11],[12]]. An effective statistical technique is the response surface methodology (RSM) which is a useful statistical tool for studying of systems where several independent variables influence the responses [[13]]. In recent years, the use of RSM in performing biological process has gained importance. The main advantage of RSM is the reduced number of tests needed to calculate multiple factors and their interactions [[14]]. In this communication, the RSM was applied to identify the suitable operating conditions for partitioning of recombinant P. fluorescens GalDH in ATPS.
Methods
Materials
Polyethylene glycols with different molecular weights were purchased from Merck (Darmstadt, Germany). d-Galactose and NAD+ were obtained from Sigma-Aldrich (St. Louis, MO, USA) and utilized in enzyme activity assay. The salts and all other chemicals were of analytical grade. P. fluorescens strain which produces GalDH enzyme has been isolated form a soil sample by Anvarsadat Kianmehr.
Construction of expression plasmid for P. fluorescens GalDH
Primers for polymerase chain reaction (PCR) amplification were designed based on the available nucleotide sequence of GalDH of the P. fluorescens genome using DNASIS MAX software (DNASIS version 3.0, Hitachi Software Engineering Co., Ltd., Tokyo, Japan). The gdh gene was amplified from the genomic DNA with specific primers GDHFw (5′-TGGATCC ATGCAACCGATTCGTCTCG-3′) and GDHRev (5′-GCGAAGCTT TTAATCGTAGAACGGC-3′), which contained the restriction sites for Bam HI and Hin dIII, respectively. PCR amplification was performed under condition: preincubation at 95°C for 1 min and then 30 cycles of 95°C for 1 min, 61°C for 1 min and 72°C for 2 min. The PCR reaction product was cut with Bam HI and Hin dIII and then ligated into the pET-28a (+) expression vector. The construct bearing the gdh gene was named pET28aGDH and transformed into E. coli BL-21 (DE3).
Cell cultivation and production of recombinant GalDH
A recombinant strain of E. coli BL21 (DE3) was grown overnight in Luria-Bertani (LB) medium containing 40 µg/mL of kanamycine at 37°C and 150 rpm. When cell density reached an OD600 of 0.8, GalDH enzyme was expressed by the addition of 0.7 mM sterile isopropyl-β-d-thiogalactopyranoside (IPTG). After 5 h of induction at 30°C, cells were harvested and stored at −20°C for further use. Pelleted E. coli cells were suspended in lysis buffer (50 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, pH 8.0), mechanically broken by sonication using a pulse sequence of 15 s on and 10 s off and clarified by centrifugation at 4,000 rpm at 4°C for 1 h. The supernatant was employed as a crude enzyme in partition experiments [[6]].
ATPS preparation
ATPS were prepared in 15-mL graduated tubes by mixing the appropriate amounts of PEG-4000, (NH4)2SO4, and enzyme solution. A final weight of 10-g system was obtained by adding a sufficient amount of 0.1 M potassium phosphate buffer (pH 8.0). Systems were agitated for 1 h at room temperature and then centrifuged at 3,000 rpm at 25°C for 40 min to speed up the phase separation. The volumes of the phases were determined, and the samples from the two phases were carefully tested for enzyme assay and total protein concentration. To avoid interference of the phase components, samples were analyzed against blanks containing the same compositions, but without enzyme [[6]]. In this work, all partition experiments were done at 25°C.
Analytical techniques
Enzyme activity was determined by monitoring the reduction of NAD+ at 340 nm. Mixture assay contained 10 mM d-galactose, 100 mM Tris-HCl buffer (pH 8.6), 2.5 mM NAD+, and the enzyme solution in a total volume of 1 mL. The change of absorbance at 340 nm was measured and corrected for blank values not including d-galactose. One unit of GalDH activity (U) is defined as the amount of enzyme catalyzing the formation of 1 μmol NADH per minute under the assay conditions [[8]]. The total protein concentration was determined by a Bio-Rad protein assay kit with bovine serum albumin (BSA) as a standard [[15]]. The purity of recombinant enzyme in ATPS was analyzed by a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Samples were diluted in a sample loading buffer and heated at 100°C for 5 min prior to being loaded into electrophoretic gel. After separation, the gel was stained with Coomassie Brilliant Blue R-250 and then destained by diffusion in a solution containing 40% (v/v) methanol and 10% (v/v) acetic acid [[16]]. The kinetic parameters of the final purified enzyme were calculated from the secondary plots of intercepts versus reciprocal concentrations of the other substrate.
Determination of partition parameters
To evaluate the partition performance of GalDH, different parameters were defined [[10]]. These include the partition coefficient (KE or KP), which is calculated as the ratio of the enzyme activity or protein concentration in the top phase divided by the correspondent value in the bottom phase. Specific activity (SA), which is defined as the enzyme activity (U/ml) in the phase sample divided by the total protein concentration (mg/ml) and is expressed in U/mg of protein.
The recovery (R%) is the ratio of the enzyme activity in the top phase (At) to the initial activity added to the system (Aori). Purification factor (PF) is calculated as the specific activity in the top phase (SAt) divided by the initial specific activity in the original sample (SAori). Yield (Y%) is determined as
where Vt and Vb are the top and bottom phase volumes, respectively.
Design of experiments and statistical analysis
A three-factor central composite face-centered design (CCFD) was used to optimize recombinant GalDH partitioning using Design-Expert software (version 8.0.4, State-Ease, Inc., USA). The selected variables were PEG-4000 concentration (X1), (NH4)2SO4 concentration (X2), and pH (X3). For each of the three variables, high (coded value +1) and low (coded value −1) points were chosen on the basis of preliminary test about their effects on GalDH partition. The level and ranges chosen for the variables are shown in Table 1. A complete CCFD experiment design allows estimation of a full quadratic model for each response. These kind of designs are easy to build because they are based on two-level factorials that have been augmented with a center point and 2k (k is the number of studied variables). Their general description is Number of experiments = 2k − p + 2k + cp, where k is the number of studied variables, p is the fractionalization element (full design, p = 0), and cp is the number of central points. All the 20 experiment (23 + 2.3 + 6) points which included six replications at the center point are described in Table 2. The experimental data obtained from the design were analyzed by the response surface regression procedure using the following second-order polynomial equation:
where Y i is the predicted response; b0, b i , b ii , and b ij are regression coefficient for the intercept, first-order model coefficients, and the linear mode coefficient for the interaction between variables i and j, respectively; and x i 's are the coded independent variables. Analysis of variance (ANOVA) was used to estimate the statistical significance of the full quadratic models. The suitability of the proposed model was evaluated by Fisher's statistical test (F test) by testing for significance between sources if variation in experimental data, i.e., the significance of the regression (SOR), the lack of fit (LOF), model p value, and the coefficient of determination (R2), results. The F value is defined as the ratio of the mean square of regression (MRR) to the error (MRe), representing the significance of each controlled variable on the tested model. The regression equations were also summated to the F test to determine the coefficient R2. The fitted polynomial equation was expressed as three-dimensional surface plots to visualize the relationship between the responses (dependent variables) and the experimental levels of each factor (independent variables) employed in the design. Parameters with less than 95% significance (p > 0.05) were removed, and the experimental data was refitted to only the significant (p < 0.05) factors to obtain the final reduced model. The combination of different optimized parameters, which gave maximum response, i.e., maximum recovery of favorite enzyme in PEG phase, was tested experimentally to confirm the validity of the model [[17],[18]].
Results and discussion
Optimization of GalDH partition process
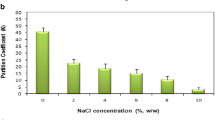
In order to elucidate the main factors that will be included in recombinant P. fluorescens GalDH partition in ATPS, a series of preliminary studies were performed. This was done in a system composed of 14% (w/w) PEG-4000 and 12% (w/w) (NH4)2SO4 ATPS at pH 7.0 and 25°C. The influence of PEG MW was investigated using four different polymer molecular weights. As shown in Figure 1, GalDH showed high affinity for the top phase. The optimal system was attained using PEG-4000, which suggested that the decreasing polymer MW until 4,000 daltons was favorable for enzyme partitioning. In contrast, the increase of PEG MW from 4,000 to 8,000 daltons resulted in less available space of GalDH in the upper phase, which led to the decrease of partition coefficient. This behavior was in agreement with an exclusion effect owing to the diminution of the free volume available in the top phase [[19]]. In accordance with the above, during examination within the framework of this research polymer, MW was not changed. To study the effect of neutral salt on the partition features of GalDH, the addition of 0% to 10% (w/w) NaCl in 14% (w/w) PEG-4000/12% (w/w) (NH4)2SO4 system was examined. Based on the obtained data (Figure 2), the highest K (45.67%) was achieved at no addition of NaCl. This result suggested that NaCl addition was not required in further modeling. The addition of salt at high concentrations leads to aggregation followed by protein precipitation, because a large amount of water molecules are strongly bound to the salts. As a consequence, the interactions among proteins become more powerful than between protein and water. Similar results for the influences of PEG MW and NaCl addition have been reported for other enzymes of oxidoreductases such as phenylalanine dehydrogenase (PheDH) [[1]] and proline dehydrogenase (ProDH) [[20]]. Meanwhile, pH range applied during this work was chosen according to the pI of target enzyme (pI = 4.86). The optimal pH range for this pI in two-phase partitioning is usually between 7.0 and 8.0 [[4]]. This was a typical behavior for enzymes which have negative charges [[21]]. The pH parameter was an important factor in process optimization, so it was included in the experimental conditions. Briefly, pH, polymer concentration, and salt concentration, which were the most important variables affecting enzyme partition were chosen for optimization in the remaining steps of this process. The optimum conditions for recombinant GalDH recovery through partitioning in PEG-ammonium sulfate ATPS were achieved by defining the experimental space around the conditions selected from preliminary tests using an experimental design methodology. The design variables and their ranges are determined as follows: PEG concentration (13% to 15%, w/w), pH (7.0 to 8.0), and salt concentration (11% to 13%, w/w). Experiments according to the design matrix of variables in Table 1 were carried out, and the relevant results are shown in Table 2. PEG and ammonium sulfate concentration as well as pH exhibited to have a significant effect on GalDH partitioning. The ANOVA was employed for the determination of significant variables and all their possible linear and quadratic interactions on the response variables. Table 3 lists the significant parameters and statistical test results of the models. The model determination coefficient, R2, was calculated to be 0.74 for SA, which indicates a good response between prediction and experimental data. The F values were calculated by the mean square (MS) of the model to the mean square of the residual error term. The model F value (10.67) and probability >F value (0.0106) for SA of recombinant GalDH indicated that the models for selected response were significant. The variables found to be statistically non-significant (p > 0.05) were removed, and the experimental data was refitted to only the significant (p < 0.05) factors to achieve the appropriate model. Larger values of F and smaller values of p showed that the variables would be significant (p < 0.05). The achieved data confirmed that the factors, pH, PEG concentration, and salt concentration, significantly affected enzyme partitioning. The response surface plots provide a method to visualize the relationship between responses and experimental levels of each variables and the type of interactions between two test variables [[22]]. Through the response surface plots, the interactions of variables and the optimum level of each variable for maximum response can be well understood. The three-dimensional graphs were generated, in which the effect of three defined factors are shown. Figure 3 depicts the response surface plots of SA against different pH values, salt concentration, and PEG concentration when one of the three variables is fixed. The individual optimum region led to a SA value (373.9 U/mg) which was achieved in the ATPS system of 14.33 % (w/w) PEG-4000 concentration, 11.79% (w/w) (NH4)2SO4 concentration, and pH 7.48. This enhancement of SA was likely the consequence of elimination of inhibitors from the optimal system which increases the enzyme activity. The highest estimated PF, Y, and R at this condition were 58.9, 92.8%, and 268.75%, respectively. As observed, the response surface plots obtained for the enzyme SA exhibited a slight increase in SA when the concentration of PEG was increased (Figure 3a,b) and a slight decline of SA as the concentration of salt was decreased (Figure 3a,c). Also, the increase of pH value resulted to reduction in enzyme SA (Figure 3b,c). The highest SA was obtained for an intermediate value of pH (7.48). This implies that there are other factors related with the characteristics of the desired enzyme (physico-chemical properties) besides the ATPS components that affect the partitioning behavior [[4]]. Collectively, it can be concluded that pH, salt concentration, and PEG concentration had a significant influence on the separation and recovery. Meanwhile, the highest values of the responses are achieved near the centers of the graph, indicating that the center points selected for this work were appropriate.
SDS-PAGE and kinetic analysis of recombinant GalDH
The partitioning of recombinant enzyme in ATPS was evaluated by SDS-PAGE analysis (Figure 4). Purified GalDH was found in the PEG-rich phase and appeared as a single protein band on Coomassie Brilliant Blue stained SDS-PAGE gel. The achieved data also demonstrated the applicability of the studied system for GalDH recovery. The subunit MW of recombinant GalDH was estimated to be about 34 kDa, which was similar to the reported value from P. fluorescens [[5],[6]]. The Km for the purified recombinant GalDH was measured to be 0.32 mM. This result showed that the obtained enzyme with ATPS had considerable affinity for its substrate and, therefore, was suitable for the diagnostic applications.
Conclusions
The work presented here showed the potential application of ATPS for partitioning and recovery of recombinant GalDH in a single step. The RSM combined to a proper factorial experimental design proved to be a powerful tool in designing and modeling the best two-phase condition for enzyme partitioning. It was concluded that the ATPS consisting 14.33% (w/w) PEG-4000 and 11.79% (w/w) (NH4)2SO4, pH 7.48 at 25°C were the most optimum system to perform GalDH partition. Under these experimental conditions, the response values for PF, R, Y, and SA were 58.9, 268.75%, 92.8%, and 373.9 U/mg, respectively, and these results were also confirmed by the evaluation of activity assay and purity of final product.
References
Shahbaz Mohammadi H, Omidinia E: Purification of recombinant phenylalanine dehydrogenase by partitioning in aqueous two-phase systems. J Chromatogr B 2007, 854: 273–278. 10.1016/j.jchromb.2007.04.049
Hatti-Kaul R: Methods in biotechnology: aqueous two-phase systems: methods and protocols. Humana Press Inc., Totowa; 1999.
Albertsson PA: Partition of cell particles and macromolecules. Wiley, New York; 1986.
Shahbaz Mohammadi H, Omidinia E: The Features of Partitioning Behavior of Recombinant Amino acid Dehydrogenase in Aqueous Two-phase Systems. In Polymer phase behavior. Edited by: Ehliers TP, Wilhelm JK. Nova Science, New York; 2011:235–264.
Sperka S, Zehelein E, Fiedler S, Fischer S, Sommer R, Buckel P: Complete nucleotide sequence of Pseudomonas fluorescens D-galactose dehydrogenase gene. Nucleic Acids Res 1989, 17: 5402–5402. 10.1093/nar/17.13.5402
Buckel P, Zehelein E: Expression of Pseudomonas fluorescens D-galactose dehydrogenase in E. coli . Gene 1981, 16: 149–159. 10.1016/0378-1119(81)90071-8
Prachayasittikul V, Ljung S, Isaraankura-Na-Ayudhya C, Bulow L: NAD (H) recycling activity of an engineered bifunctional enzyme galactose dehydrogenase/lactate dehydrogenase. Int J Biol Sci 2006, 2: 10–16. 10.7150/ijbs.2.10
Mazitsos BA, Rigden DJ, Tsoungas PG, Clonis YD: Galactosyl-mimodye ligands for Pseudomonas fluorescens β-galactose dehydrogenase. Eur J Biochem 2002, 269: 5391–5405. 10.1046/j.1432-1033.2002.03211.x
Fughmura Y, Kawamura M, Naruse H: A new method of blood galactose estimation for mass screening of galactosomia. Tohoku J Exp Med 1981, 133: 371–378. 10.1620/tjem.133.371
Teles de Faria J, Coelho Sampaio F, Converti A, Lopes Passos FM, Paula Radrigues Minim V, Antonio Minim L: Use of response surface methodology to evaluate the extraction of Debaryomyces hansenii xylose reductase by aqueous two-phase system. J Chromatogr B 2009, 877: 3031–3037. 10.1016/j.jchromb.2009.07.023
Dembczynski R, Białas W, Jankowski T: Partitioning of lysozyme in aqueous two-phase systems containing ethylene oxide-propylene oxide copolymer and potassium phosphates. Food Bioprod Process 2013, 91: 292–302. 10.1016/j.fbp.2012.11.001
Garai D, Kumar V: Aqueous two phase extraction of alkaline fungal xylanase in PEG/phosphate system: optimization by Box–Behnken design approach. Biocatal Agric Biotechnol 2013, 2: 125–131.
Zhu W, Song J, Ouyang F, Bi J: Application of response surface methodology to the modeling of α–amylase purification by aqueous two-phase systems. J Biotechnol 2005, 118: 157–165. 10.1016/j.jbiotec.2005.03.017
Singh P, Singh Shera S, Banik J, Mohan Banik R: Optimization of cultural conditions using response surface methodology versus artificial neural network and modeling of L-glutaminase production by Bacillus cereus MTCC 1305. Bioresour Technol 2013, 137: 261–269. 10.1016/j.biortech.2013.03.086
Bradford MM: Rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principles of protein-dye binding. Anal Biochem 1976, 72: 248–254. 10.1016/0003-2697(76)90527-3
Sambrook J, Fritsch EF, Maniatis T: Molecular cloning: a laboratory manual, 2nd edn.. Cold Spring Harbor Laboratory press, Cold Spring Harbor; 1994.
Treier K, Lester P, Hubbuch J: Application of genetic algorithms and response surface analysis for the optimization of batch chromatographic systems. Biochem Eng J 2012, 63: 66–75. 10.1016/j.bej.2012.02.003
Xu Q, Shen Y, Wang H, Zhang N, Xu S, Zhang L: Application of response surface methodology to optimize extraction of flavonoids from fructus sophorae. Food Chem 2013, 138: 2122–2129. 10.1016/j.foodchem.2012.11.099
Yücekan I, Onal S: Partitioning of invertase from tomato in poly (ethylene glycol)/sodium sulfate aqueous two-phase systems. Process Biochem 2011, 40: 226–232. 10.1016/j.procbio.2010.08.015
Shahbaz Mohammadi H, Omidinia E: Process integration for the recovery and purification of recombinant Pseudomonas fluorescens proline dehydrogenase using aqueous two-phase systems. J Chromatogr B 2013, 929: 11–17. 10.1016/j.jchromb.2013.03.024
Yan-Min L, Yan-Zaho Y, Xi-Dan Z, Chuan-Bo X: Bovine serum albumin partitioning in polyethylene glycol (PEG)/potassium citrate aqueous two-phase systems. Food Bioprod Process 2010, 88: 40–46. 10.1016/j.fbp.2009.12.002
Kammoun R, Chouyekh H, Abid H, Naili B, Bejar S: Purification of CBS 819.72 α-amylase by aqueous two-phase systems: modeling using response surface methodology. Biochem Eng J 2009, 46: 306–312. 10.1016/j.bej.2009.06.003
Acknowledgements
We would like to express our thanks to Dr. Hamid Shahbaz Mohammadi for revising the text.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AK carried out the design of experiments with RSM and purification and characterization of the target enzyme. MP did the general molecular biology methodology in this research. BM participated in the microbiology sections and also the paper writing. SS participated in the coordination of the manuscript as well as doing the statistical analysis. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kianmehr, A., Pooraskari, M., Mousavikoodehi, B. et al. Recombinant d-galactose dehydrogenase partitioning in aqueous two-phase systems: effect of pH and concentration of PEG and ammonium sulfate. Bioresour. Bioprocess. 1, 6 (2014). https://doi.org/10.1186/s40643-014-0006-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40643-014-0006-8