Abstract
Nonribosomal peptides are products that fall into the class of secondary metabolites with a diverse properties as toxins, siderophores, pigments, or antibiotics, among others. Unlike other proteins, its biosynthesis is independent of ribosomal machinery. Nonribosomal peptides are synthesized on large nonribosomal peptide synthetase (NRPS) enzyme complexes. NRPSs are defined as multimodular enzymes, consisting of repeated modules. The NRPS enzymes are at operons and their regulation can be positive or negative at transcriptional or post-translational level. The presence of NRPS enzymes has been reported in the three domains of life, being prevalent in bacteria. Nonribosomal peptides are use in human medicine, crop protection, or environment restoration; and their use as commercial products has been approved by the U. S Food and Drug Administration (FDA) and the U. S. Environmental Protection Agency (EPA). The key features of nonribosomal peptides and NRPS enzymes, and some of their applications in industry are summarized.
Similar content being viewed by others
Background
Nonribosomal peptides is a diverse family of natural products fall into the class of secondary metabolites with a diverse properties as toxins, siderophores, pigments, antibiotics, cytostatics, immunosuppressants or anticancer agents [1, 2]; and have a particularity: their synthesis is independent of ribosomal machinery. Soil-inhabiting microorganisms, such as Actinomycetes and Bacilli, and eukaryotic filamentous fungi are mostly producers of nonribosomal peptides, but marine microorganisms have also emerged as a source for such peptides [3]. These peptides have a structural features such as contain amino acids like ornithine or imino acids, and their structures are macrocyclic, branched macrocyclic, dimers or trimers of identical structural elements [4]. Usually nonribosomal peptides are synthesized on large nonribosomal peptide synthetase (NRPS) enzyme complexes, defined as modular multidomain enzymes; nevertheless more than half of the NRPS enzymes found in a genome-mining study of 2699 genomes by Wang et al. [1] are nonmodular NRPS enzymes. Nonmodular NRPS enzymes are found in siderophore biosynthetic pathways like EntE and VibH in enterobactin, and VibE in vibriobactin [5] or as a stand-alone peptidyl carrier protein such as BlmI from the bleomycin gene cluster [6]. The presence of NRPS enzymes has been reported in the three domains of life, being prevalent in bacteria, less frequent in eukarya and rare in archaea. Within bacteria domain, Proteobacteria, Actinobacteria, Firmicutes, and Cyanobacteria were the phyla with major abundance of these enzymes, and there has been observed a correlation between genome size and the number of NRPS’s clusters [1]. The key features of the microbial biosynthesis of nonribosomal peptides, the structure and regulation of NRPS enzymes, and some applications in industry are summarized below.
Biosynthesis of nonribosomal peptides
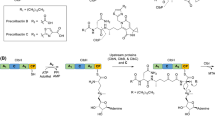
The biosynthesis of nonribosomal peptides is done by NRPSs which are modularly organized multi-enzyme complexes which serve as templates and biosynthetic machinery, via a thiotemplate mechanism independent of ribosomes [2]. A module is defined as a section of the NRPS enzyme that incorporate in a specific manner one amino acid into the peptide backbone, and in turn the modules can be divided into domains, which catalyze the individual steps of nonribosomal peptide synthesis. Each module consist of three domains, adenylation (A) domain, peptidyl carrier protein (PCP) or thiolation (T) domain, and condensation (C) domain which carry out the synthesis of nonribosomal peptides (Fig. 1) [7]. The order of modules is usually co-linear to the product peptide sequences [8, 9]. Nonribosomal peptides synthesis proceeds in a N- to C-terminal direction, producing peptides that are usually about 3–15 amino acids in length, and the released peptides can be linear, cyclic, or branched-cyclic [3]. There is a sequence motif conserved in domains that facilitates the identification of these using sequences search tools like BLAST, as in the study done by Etchegaray et al. [10] to identified NRPSs in the genome of Xanthomonas axonopodis and X. campestri. The first step in the biosynthesis is done by the A-domain, which recognizes and performs activation of amino acid substrate via adenylation using Mg-ATP, result in an aminoacyl adenylated intermediate [11]. The A-domain consist of ~550 amino acids, and has ten amino acid residues that can be considered as the “codons” of NRPS enzymes and are important to substrate specificity [12]. The substrates that can be recognized by A-domain can include D and L forms of the 20 amino acids used in ribosomal proteins synthesis, as well as non-proteino-genic amino acids such as ornithine, imino acids, and hydroxy acids such as α-aminoadipic and β-butyric acids [2]. The reaction carried out by the A-domain shares the same chemistry as that performed by aminoacyl-tRNA synthetases (AARSs), which act in the initial step of ribosome-driven protein synthesis [8, 13]. The second step is performs by the PCP-domain of ~80 amino acids [12], which covalently binds the activated amino acid to their cofactor 4′-phosphopantetheine (PP) arm via a thioester bond and transfer the activated substrate and the elongation intermediates to C-domain [14]. The final step is carried out by the C-domain of ~450 amino acids, which catalyzes peptide bond formation between the carboxyl group of the nascent peptide and the amino acid carried by the flanking module, allowing the translocation of the growing chain to the following module [12, 13]. After condensation step, the linear intermediate peptide is release with the help of thioesterase (TE) domain by either hydrolysis or internal cyclization in bacterias, and less frequent in NRPSs of fungi. In fungi the chain release is carried out by a variety of mechanisms, two of which are (1) a terminal C domain, which catalyzes release by inter- or intra-molecular amide bond formation, and (2) a thioesterase NADP(H) dependent reductase (R) domain which catalyzes reduction with NADPH to form an aldehyde [2]. The primary product of this synthesis may be post-synthetically modified to achieve its mature form by additional tailoring enzymes which are not part of NRPS by N-, C- and O-methylation, glycosylation, hydroxylation, acylation, halogenation or heterocyclic ring formation [3, 15, 16]. These tailoring enzymes and their reactions contribute to generate the structural diversity of nonribosomal peptides [10].
Structural features of nonribosomal peptide synthetase enzymes. Nonribosomal peptide synthetase enzymes can be subdivided in modules, each incorporating one amino acid. Each module consist of three domains: adenylation (A) domain, peptidyl carrier protein (PCP) domain, and condensation (C) domain, which carry out the synthesis of nonribosomal peptides; the epimerization (E) domain and thioesterase (TE) domain is also represents. In this figure, surfactin biosynthesis was exemplified
Structure of nonribosomal peptide synthetase (NRPS) enzymes
NRPSs are defined as multimodular enzymes, consisting of repeated modules with A-PCP-C domains [17]. Multimodular NRPS proteins are frequently in fungal genomes [3], and all modules that participate in the assembly of a peptide are connected within a single enzyme [10]. Tandem duplication and recombination is may be the origin of modules of multimodular NRPS enzymes, as in the case of SimA (Cyclosporin synthetase) enzyme of Tolypocladium inflatum and Enniatin synthetase of Fusarium equiseti, respectively [2]. Nevertheless, more than half of the NRPS enzymes finding in a genome-mining study of 2699 genomes are nonmodular NRPS enzymes, being common in bacterial systems where they are organized in clusters [1, 3, 10]. Nonmodular NRPS enzymes consisting of one or two A-PCP-C modules, or lacked complete A-PCP-C modules and consist of a single A-domain or an A-PCP unit followed by a variety of C-terminal domains [2]. In bacterial systems, mono or bi modular NRPSs can interact with others NRPS proteins and performs biosynthesis by first activating the amino acid and then transferred the activated substrate either to a C domain in the same NRPS or in a different NRPS, known as nonlinear biosynthesis [3]. Taxonomic distributions of mono/bi modular NRPS subfamilies suggest an ancient origin, possibly predating the divergence of eubacteria and fungi [2]. There is third structure of NRPSs which is fused to a polyketide synthase (PKS) unit, both types of synthetases are fused in a single polypeptide [10]. Of 3339 gene clusters encoding NRPS and PKS found by Wang et al. [1], one-third (1147) of gene clusters belonged to the hybrid type and encoded 462 hybrid proteins that contain both NRPS and PKS core domains. PKSs are large megasynthases related to fatty acid synthases, that biosynthesize small molecule polyketides with diverse natural function as nonribosomal peptides [2].
Regulation of NRPS genes
Global regulators such as DegU and Comp/ComA two-component system can act at transcriptional level, regulating positively the expression of srfA, bac, and bmy cluster genes of Bacillus spp., which encodes surfactin (Fig. 1), bacilysin and bacillomycin, respectively [18–20]. In the case of srfA genes that encoding for the surfactin, the transcriptional initiation is through ComX pheromone that can activated ComP, causing ComP to autophosphorylate and, subsequently, donate a phosphate group to ComA. Upstream of the srfA promoter there are ComA boxes, which are recognized by phosphorylated ComA and binding to them as a tetramer initializing the transcription of srfA [18, 19, 21]. Another positive regulator of transcription of srfA is PerR protein, which positively modulates srfA expression by binding to regions located upstream of ComA boxes, known as PerR boxes [22]. Expression of the bac operon is dependent on a σA-dependent promoter, which is activated by interaction with DegU at the final stage of vegetative growth. Binding of DegU to the bac operon promoter occurred mainly at three sites, in a similar way that occur with bmy operon, in which DegU binds directly to two sites located upstream of the bacillomycin D promoter [20, 23]. In the opposite case, a down-regulation is exerted on the bac operon through ScoC protein, which binds at the bac promoter sequence in ScoC boxes located between positions −50 and −42 (ScoC box1) and between positions −12 and −4 (ScoC box2) [24]. While srfA is repressed by CodY at high concentrations of amino acids Ile, Leu, and Val. AbrB and Spx are two other negative regulators that turn off the transcription of srfA genes [19]. In the case of SrfA NRPS enzyme, is necessary a post-translational modification to become active; 4′-phosphopantetheinyl transferase (Sfp/PPTase) is required for the activation of SrfA enzymes by converting the inactive apo-forms to active holo-forms [25]. In the case of Streptomyces peucetius, the biosynthesis of non-ribosomal peptide doxorubicin, an antitumor drugs, is up regulated by the networking of transcriptional regulators dnrO, dnrN, and dnrI. The product of dnrO gene binds to the dnrN/dnrO promoter region and activates dnrN. DnrN activates the transcription of dnrI gene and, DnrI activates the transcription of the doxorubicin biosynthesis genes. Down regulation of doxorubicin is indirected controlled by doxR regulator, a gene belonging to the IclR family of transcripional regulators, which inhibits the dnrI expression, leading to blockade of doxorubicin production [26].
Environmental applications
Surfactants are amphipathic molecules with both hydrophilic and hydrophobic moieties. These properties render surfactants capable of reducing surface and interfacial tension and forming microemulsion. Such characteristics confer excellent detergency, emulsifying, foaming, and dispersing traits; they are the active ingredients found in soaps and detergents [27]. Surfactants currently in use are chemically derived from petroleum; however, interest in microbial surfactants has been steadily increasing in recent years due to their diversity, environmentally friendly nature, and their potential applications in the environmental protection, crude oil recovery, health care, and food-processing industries [28–30]. Biosurfactants are biological surface-active compounds largely produced by a wide variety of microorganisms, secreted either extracellularly or attached to parts of cells, predominantly during growth on water-immiscible substrates. Bacteria of various genera such as Pseudomonas, Bacillus, Acinetobacter, Arthrobacter, and Rhodococcus are able to produce biosurfactants during hydrocarbon oxidation [31]. Biosurfactants have several properties and advantages over the chemical surfactants, such as lower toxicity, higher biodegradability, better environmental compatibility, higher foaming, high selectivity and specific activity at extreme temperatures, pH and salinity [28]. Biosurfactants are capable of lowering surface and interfacial tensions effectively and thus are potential substitutes for widely used chemically synthesized surfactants, they have very low critical micelle concentrations (CMC), this means that are effective at low concentrations. In general, the structure of these molecules includes a hydrophobic portion commonly made up of fatty acids (saturated, unsaturated, or hydroxylated), whereas the hydrophilic portion is usually composed of peptides or mono-, di-, or polysaccharides [26]. The major classes of biosurfactants include glycolipids, lipopeptides, lipoproteins, phospholipids, polysaccharide-lipid complexes, hydroxylated and cross-linked fatty acids, and the complete cell surface [27]. One of the potential uses is in the oil industry, in which case whole-cell broth could be used with minimum purity specification and required in small quantities to oil recovery from underground sandstone. Another use of biosurfactants is in remediation of hydrocarbon and crude oil-contaminated soils (Fig. 2b); the addition of biosurfactant increase the bioavailability of petroleum hydrocarbon pollutants in soil to stimulate the indigenous bacterial population to degrade hydrocarbons at rates higher [32]. One example is the use of rhamnolipid biosurfactant from Pseudomona aeruginosa that removed oil from contaminated Alaskan gravel from the Exxon Valdez oil spill [33]. From studies conducted by Urum and Pekdemir [27], it is noted that biosurfactants were able to remove significant amount of crude oil from contaminated soil, for instance rhamnolipid removed up to 80 % oil and lecithin about 42 %. While in the studies done by Lai et al. [32], was shown that rhamnolipid from P. aeruginosa and surfactin from B. subtillis have a higher removal efficiency from a heavy oil-polluted site, than the chemicals surfactants Tween 80 and triton X-100. In the case of surfactin, has been reported that its production has reached concentrations of 10.26 g/L in a medium containing starch [34] 2933 times higher than that achieved by one report by Ponte Rocha et al. [35]. Rhamnolipid production using P. aeruginosa mutant strains grown in blackstrap molasses with or without supplementary nitrogen source was of 1.45 g/L after 96 h incubation [36]. While Silva et al. [37] using P. aeruginosa UCP0992 grown in 100 ml mineral medium (aeration of 80 %) supplemented with 3 % glycerol and 0.6 % NaNO3, as the nitrogen source, at 28 °C after 96 h reached a production of 8.0 g/L; similarly Wu et al. [38] using an indigenous strain P. aeruginosa EM1 originating from an oil-contaminated site located in southern Taiwan grown in inorganic nitrogen (NaNO3) obtained a productivity of 8.63 g/L. To avoid the inconvenience of working with opportunistic pathogen strains like P. aeruginosa, attempts have been made to express biosurfactants using non-pathogenic strains of bacteria. Wittgens et al. [39] using heterologous expression in Pseudomonas putida KT42C1, a strain certified as safety, produced up to 1.5 g/L of rhamnolipid; while Ochsner et al. [40] reached a production of 0.6 g/L in a recombinant P. putida strain KT2442. Escherichia coli has also been used to production of biosurfactants, as the case reported by Wang et al. [41], with the engineered E. coli TnERAB that produced 65–80 mg/L in MS plus glucose media, and 150–185 mg/L in LB plus glucose media, respectively.
Human health applications
Since the discovery of penicillin in 1928 by Alexander Fleming to 1940, the efforts to produce penicillin have conducted the biotechnology sector into a billion dollar industry with deep-tank fermentations at its core [42]. The fungi Penicillium chrysogenum is the organism utilized to produce penicillin at industrial scale. Penicillins are formed from the amino acids valine, cysteine and α-aminoadiapate and include residues such as pheniylacetyl [43]. The penicillin biosynthetic pathway encompasses δ-(L-α-aminoadipyl)-L-cysteinyl-d-valine synthetase (ACVS), isopenicillin N synthase (IPNS), isopenicillin N acyl transferase (IAT) and phenylacetyl CoA ligase (PCL), with the ACVS belonging to a class of NRPSs that exclusively occurs in certain filamentous fungi and bacteria (Actinomycetes, Bacilli) [44]. The production of penicillin have been reported in bioreactors of 100,000 L, achieving 36 g/L at 250 h [42], although 50 g/L of penicillin can be produced [45]. Therefore, the penicillin fermentation process is a good case of a development strategy model to follow into a large scale of nonribosomal peptide process production. However, the residual concentrations obtained of many NRP are from one to three orders of magnitude below compared to that of penicillin. For example, the cyclic undecapeptide Cyclosporin A is synthetized by cyclosporine synthetase one of the most complex and largest modular enzymes described [46]. Cylcosporine A is produced by fungus Tolypocladium inflatum, Beauveria nivea, Fusarium roseum, and Tolypocladium niveum and have anti-inflammatory, inmunosupressive, antifungal and antiparasitic properties [47]. Maximum Cyclosporin A production of 1274 mg/L with Tolypocladium inflatum was reported by Survase et al. [48] in submerged fermentation. In case of the Echinocandins, novel antymicotics produced by ascomycota fungi have a cyclic lipo-hexapeptide structure and act as β-1,3-glucan synthase inhibitors [49]. Echinocandin B the precursor of anidulafungin, is produced by Aspergillus nidulans and had reached 1.5 g/L in potato dextrose broth (PDB) [50]. The pneumocandin B0 precursor of caspofungin have reached about 2 g/L in the fermentation broth of Glarea lozoyensis [51]. Kanda et al. [52] reported the screening of a mutant of Coleophoma empetri and improved medium conditions for production of the antibiotic FR901379, the precursor of micafungin. The mutant strain had a 30-fold higher productivity compared to the wild type which produced 1 U/mL. In 2010, Kanda et al. [52] demonstrated the production of FR901379 with optimal conditions in a fermenter of 15,000 l reaching 50 U/mL. In the other hand, the bacteria Actinomycetes are known for produce novel bioactive compounds; more than 10,000 compounds were described only from genus Streptomyces [53]. Many of these compounds are synthesized by polyketide synthases (PKSs). In case of NRP, bleomycin is a glycopeptide produced by Streptomyces verticillus with antibacterial and antitumor properties [54] and is produced at 10 mg/L [55]. Daptomycin is a lipopeptide produced by Streptomyces roseosporus and consist of 13 aminoacids and have been approved because is effective for treatment of skin and skin structure infections caused by gram-positive pathogens [56]. A production of 812 mg/L has been reached through fed-batch fermentation with feedback control of dextrin [57]. In case of the peptide antibiotics, bacitracins (Fig. 2a, b) are produced by some species of Bacillus licheniformis and Bacillus subtilis, in addition contains at least 10 distinct dodecapeptides that differ by one or two aminoacids [58]. The bacitracin A production has reached approximately 900 mg/L in cultures of B. licheniformis NCIMB 8874 with the addition of oligosaccharides as elicitor [59]. Polymixns consist of ten amino acids with a characteristic polycationic heptapeptide ring and an N-terminal fatty acid modification and are produced by B. subtilis and Peanibacillus polymyxa [60]. Although this antibiotic is commercial, information about its level of concentration reached in production is vague. The issues of toxicity of certain NRPs have been described. For example, the family of polymixins such as polymyxin B and polymyxin E (colistin) that are cyclic lipopeptides produced by P. polymyxa were introduced into clinical medicine in the late 1950s but its use waned in the 1970s because the adverse effects in nephrotoxicity [61–63]. Nonetheless, its use has recently increased because the colistin is one of the antibiotics used for multidrug-resistant infections of Gram-negative bacteria such as P. aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae [61, 64]. Schneditz et al. [65] reported that the NRP tilivalline produced by Klebsiella oxytoca have pathophysiological effects on human epithelial cells through the induction of apoptosis and disruption of epithelial barrier function. Because of the toxicity that can present some NRPs such as lipopeptides, efforts have been made to reduce cytotoxicity. For example, Jiang and colleagues [66] develop a novel class of antibacterial lipopeptides from surfactin that have a reduced cytotoxicity with no significant diminution in antimicrobial activity; another case is the mentioned by Robbel and Marahiel [67] with the daptomycin, a branched cyclic lipopeptide antibiotic, which was subjected to a deacylation to reduce its cytotoxicity.
Commercial and market opportunities for NRPs
Due to their environment-friendly properties, low toxicity and higher biodegradability, the NRPs such as biosurfactants have had an increase in their demand for use in biotechnological applications, for example, in extraction of petroleum, environmental restoration, foods, beverages, cosmetics, detergents or medicine [68, 69]. The total quantity of chemical and biological surfactants produced in the US is estimated at more than 10 billion pounds, and worldwide at 25 billion pounds [70]. Regarding only to biosurfactants, the reports for the values of the world market and its poduction in tonnes varies. Reis et al. [71] reports that the global market had a value of USD 1.7 billion in 2011 and is expected to reach USD 2.2 billion in 2018, based on a growth rate of 3.5 % per annum, and the global biosurfactants market volume is expected to reach 476,512.2 tons by 2018. While Campos et al. [72] reports that revenues from the biosurfactants market were USD 6.5 billion in 2012, and the market volume was 3.5 million tons. But regardless of the variation in the reports of market value, both the volume and the value of the biosurfantes is increasing. Currently the cost of biosurfactants in the market is high, compared with chemical surfactants. For example, surfactin (98 % purity) from B. subtilis available from Sigma Chemical Company has a cost of $191 for a 10 mg vial, while the cost of the rhamnolipids (95 % purity) from P. aeruginosa is $379 for 10 mg vial; in contrast, chemical surfactants as Alkanol® XC (Sigma Chemical Company), have a market cost of $72 for 500 g. Although at first glance the cost of chemical surfactants is much lower than the biosurfactants, due to environmental damage that they can cause, eventually their cost is much higher. The use of expensive substrates, limited product concentrations, low yields and formation of product mixture rather than pure compounds, are some reasons for limited use microbial surfactants and their high cost [73, 74]. Despite its high costs, biodegradability and low ecotoxicity of biosurfactants are features that draw the attention of companies as Ecover, which is a Belgian manufacturer of ecological detergents and cleansing agents, which use the biosurfactants sophorolipids in hard surface cleaners such as multisurface cleaner, floor soap, and window cleaner; or the Japanese company Saraya Co. LTD, which commercialized a dish washer containing sophorolipids as cleaning agent. Sophorolipids are also used in cosmetics products, for example, the French company Soliance produces sophorolipid-based cosmetics for body and skin; the Korean MG Intobio Co. Ltd commercializes Sopholine cosmetics, which is functional soap specific for acne treatment; or the Japanese company Kao Co. Ltd., which uses sophorolipids as humectants for cosmetic makeup brands such as Sofina [75, 76]. Biosurfactants also have been applied in the food industry, for example, rhamnolipids can be found as active substance in the fungicide Zonix™, produced by the company Jeneil Biotech Inc and approved by FDA for use on vegetables, legumes, and fruits crops [77, 78]. In the medical field there are examples of use of NRPs, such as Cyclosporin A and bleomycin A2. Cyclosporine A is an immunosuppressive agent, which has its application in the aftercare of organ transplantations; while bleomycin A2 exhibit cytostatic activity, which makes it suitable for cancer therapy [79]. These NRPs have high selling prices in the market. The cost of these molecules available from Sigma Chemical Company is $107 for 25 mg of Cyclosporin A (98 % purity) extracted from T. inflatum, and $847 for 20 mg of bleomycin A2 (70 % purity) extracted from S. verticillus. The use of biosurfactants increases, as well as investigations that result in patents for commercial use [77], but still need to reduce their production costs in order to be competent in terms of their prices.
Conclusions
Regarding the information of some commercial NRP described above, the concentration reached in production of them is still poor. Many studies have been made of production of NRP that are candidates to be utilized in different applications. However, its production also is deficient. Hence, the bioprocess engineering approaches must work together with the approach of bioinformatics genome mining, the heterologous production of NRP, the improvement of biosynthetic pathways as well as the physiology of the producer cells. Taken examples like process development penicillin, we will capable to produce many NRPs in larger quantities in order to overcome many problems in human health, crop protection, food industry as well as in environmental applications.
References
Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K (2014) Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci 111(25):9259–9264
Bushley KE, Turgeon BG (2010) Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol 10:26
Mootz HD, Schwarzer D, Marahiel MA (2002) Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem 3(6):490–504
Singh RK, Singh P, Mohapatra TM (2012). Nonribosomal peptide synthesis in microbes. In Recent advances in microbiology. Nova Science Publishers, Inc, New York
Crosa JH, Walsh CT (2002) Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66(2):223–249
Du L, Shen B (1999) Identification and characterization of a type II peptidyl carrier protein from the bleomycin producer Streptomyces verticillus ATCC 15003. Chem Biol 6(8):507–517
Drake EJ, Miller BR, Shi C, Tarrasch JT, Sundlov JA, Allen CL, Skiniotis G, Aldrich CC, Gulick AM (2016) Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature 529(7585):235–238
Finking R, Marahiel MA (2004) Biosynthesis of nonribosomal peptides1. Annu Rev Microbiol 58:453–488
Hahn M, Stachelhaus T (2004) Selective interaction between nonribosomal peptide synthetases is facilitated by short communication-mediating domains. Proc Natl Acad Sci 101(44):15585–15590
Etchegaray A, Silva-Stenico ME, Moon DH, Tsai SM (2004) In silico analysis of nonribosomal peptide synthetases of Xanthomonas axonopodis pv. citri: identification of putative siderophore and lipopeptide biosynthetic genes. Microbiol Res 159(4):425–437
Miller BR, Sundlov JA, Drake EJ, Makin TA, Gulick AM (2014) Analysis of the linker region joining the adenylation and carrier protein domains of the modular nonribosomal peptide synthetases. Proteins 82(10):2691–2702
Tanovic A, Samel SA, Essen LO, Marahiel MA (2008) Crystal structure of the termination module of a nonribosomal peptide synthetase. Science 321(5889):659–663
Tiburzi F, Visca P, Imperi F (2007) Do nonribosomal peptide synthetases occur in higher eukaryotes? IUBMB Life 59(11):730–733
Reimer JM, Aloise MN, Harrison PM, Schmeing TM (2016) Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase. Nature 529(7585):239–242
Walsh CT, Chen H, Keating TA, Hubbard BK, Losey HC, Luo L, Marshall CG, Miller DA, Patel HM (2001) Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr Opin Chem Biol 5(5):525–534
Losey HC, Peczuh MW, Chen Z, Eggert US, Dong SD, Pelczer I, Kahne D, Walsh CT (2001) Tandem action of glycosyltransferases in the maturation of vancomycin and teicoplanin aglycones: novel glycopeptides. Biochemistry 40(15):4745–4755
Weissman KJ (2015) The structural biology of biosynthetic megaenzymes. Nat Chem Biol 11(9):660–670
Wang X, Luo C, Liu Y, Nie Y, Liu Y, Zhang R, Chen Z (2010) Three non-aspartate amino acid mutations in the ComA response regulator receiver motif severely decrease surfactin production, competence development and spore formation in Bacillus subtilis. J Microbiol Biotechnol 20(2):301–310
Roongsawang N, Washio K, Morikawa M (2010) Diversity of nonribosomal peptide synthetases involved in the biosynthesis of lipopeptide biosurfactants. Int J Mol Sci 12(1):141–172
Mariappan A, Makarewicz O, Chen XH, Borriss R (2012) Two-component response regulator DegU controls the expression of bacilysin in plant-growth-promoting bacterium Bacillus amyloliquefaciens FZB42. J Mol Microbiol Biotechnol 22(2):114–125
Nakano MM, Zuber P (1993) Mutational analysis of the regulatory region of the srfA operon in Bacillus subtilis. J Bacteriol 175(10):3188–3191
Hayashi K, Ohsawa T, Kobayashi K, Ogasawara N, Ogura M (2005) The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis. J Bacteriol 187(19):6659–6667
Koumoutsi A, Chen XH, Vater J, Borriss R (2007) DegU and YczE positively regulate the synthesis of bacillomycin D by Bacillus amyloliquefaciens strain FZB42. Appl Environ Microbiol 73(21):6953–6964
Inaoka T, Wang G, Ochi K (2009) ScoC regulates bacilysin production at the transcription level in Bacillus subtilis. J Bacteriol 191(23):7367–7371
Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT (1998) Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37(6):1585–1595
Chaudhary AK, Singh B, Maharjan S, Jha AK, Kim BG, Sohng JK (2014) Switching antibiotics production on and off in actinomycetes by an IclR family transcriptional regulator from Streptomyces peucetius ATCC 27952. J Microbiol Biotechnol 24(8):1065–1072
Urum K, Pekdemir T (2004) Evaluation of biosurfactants for crude oil contaminated soil washing. Chemosphere 57(9):1139–1150
Desai JD, Banat IM (1997) Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev 61(1):47–64
Marchant R, Banat IM (2012) Biosurfactants: a sustainable replacement for chemical surfactants? Biotechnol Lett 34(9):1597–1605
Jackson SA, Borchert E, O’Gara F, Dobson ADW (2015) Metagenomics for the discovery of novel biosurfactants of environmental interest from marine ecosystems. Curr Opin Biotechnol 33:176–182
Kuyukina MS, Ivshina IB, Makarov SO, Litvinenko LV, Cunningham CJ, Philp JC (2005) Effect of biosurfactants on crude oil desorption and mobilization in a soil system. Environ Int 31(2):155–161
Lai CC, Huang YC, Wei YH, Chang JS (2009) Biosurfactant-enhanced removal of total petroleum hydrocarbons from contaminated soil. J Hazard Mater 167(1–3):609–614
Harvey S, Elashvili I, Valdes JJ, Kamely D, Chakrabarty AM (1990) Enhanced removal of Exxon Valdez spilled oil from Alaskan gravel by a microbial surfactant. Biotechnology 8(3):228–230
Gong G, Zheng Z, Chen H, Yuan C, Wang P, Yao L, Yu Z (2009) Enhanced production of surfactin by Bacillus subtilis E8 mutant obtained by ion beam implantation. Food Technol Biotechnol. 47(1):27–31
Ponte Rocha MV, Gomes Barreto RV, Melo VMM, Barros Goncalves LR (2009) Evaluation of cashew apple juice for surfactin production by Bacillus subtilis LAMI008. Appl Biochem Biotechnol 155(1–3):366–378
Raza ZA, Khan MS, Khalid ZM (2007) Physicochemical and surface-active properties of biosurfactant produced using molasses by a Pseudomonas aeruginosa mutant. J Environ Sci Health A Tox Hazard Subst Environ Eng. 42(1):73–80
Silva SN, Farias CB, Rufino RD, Luna JM, Sarubbo LA (2010) Glycerol as substrate for the production of biosurfactant by Pseudomonas aeruginosa UCP0992. Colloids Surf B Biointerfaces. 79(1):174–183
Wu JY, Yeh KL, Lu WB, Lin CL, Chang JS (2008) Rhamnolipid production with indigenous Pseudomonas aeruginosa EM1 isolated from oil-contaminated site. Bioresour Technol 99(5):1157–1164
Wittgens A, Tiso T, Arndt TT, Wenk P, Hemmerich J, Müller C, Wichmann R, Küpper B, Zwick M, Wilhelm S, Hausmann R, Syldatk C, Rosenau F, Blank LM (2011) Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb Cell Fact 10:80
Ochsner UA, Reiser J, Fiechter A, Witholt B (1995) Production of Pseudomonas aeruginosa rhamnolipid biosurfactants in heterologous hosts. Appl Environ Microbiol 61(9):3503–3506
Wang Q, Fang X, Bai B, Liang X, Shuler PJ, Goddard WA 3rd, Tang Y (2007) Engineering bacteria for production of rhamnolipid as an agent for enhanced oil recovery. Biotechnol Bioeng 98(4):842–853
Goldrick S, Ştefan A, Lovett D, Montague G, Barry Lennox (2015) The development of an industrial-scale fed-batch fermentation simulation. J Biotechnol 193:70–82
PrauBe MTE, Schäuble S, Guthke R, Schuster S (2016) Computing the various pathways of penicillin synthesis and their molar yields. Biotechnol Bioeng 113:173–181
Gidijal L, Kiel JAKW, Douma RD, Seifar RM, van Gulik WM, Bovenberg RAL, Veenhuis M, Van der Klei IJ (2009) An engineered yeast efficiently secreting penicillin. PLoS ONE 4(12):e8317
Jørgensen H, Nielsen J, Villadsen J, Møllgaard H (1995) Meatbolic flux distributions in Penicillum chrysogenum during fed-batch cultivations. Biotechnol Bioeng 46:117–131
Lawen A, Zocher R (1990) Cyclosporin Synthetase J Biol Chem 265:11355–11360
Survase SA, Kagliwal LD, Annapure US, Singhal RS (2011) Cyclosporin A—a review on fermentative production, downstream processing and pharmacological applications. Biotechnol Adv 29:418–435
Survase S, Bankar SB, Annapure AS, Singhal RS (2015) The effect of agitation and aeration on production of cylcosporinA n batch cultures of Tolypocladium inflatum. Indian J Biotechnol 14(394):401
Emri T, Majoros L, Tóth V, Pócsi I (2013) Echinocandins: production and applications. Appl Microbiol Biotechnol 97:3267–3284
Zou SP, Liu M, Wang QL, Xiong Y, Niu K, Zheng YG, Shen YC (2015) Preparative separation of echinocandin B from Aspergillus nidulans broth using macroporous resin adsorption chromatography. J Chromatography B 978–979:111–117
Balkovec JM, Hughes DL, Masurekar PS, Sable CA, Shwartz RE, Singh SB (2014) Discovery and development of first in class antifungal caspofungin (CANCIDAS®)-A case of study. Nat Prod Rep 31:15–34
Kanda M, Tsuboi M, Sakamoto K, Shimizu S, Yamashita M, Honda H (2009) Improvement of FR901379 production by mutant selection and medium optimization. J Biosc Bioeng 107:530–534
Bérdy J (2012) Thoughts and facts about antibiotics: where are now and where are heading. J Antib 65(385):395
Gu J, Codd R (2012) Copper(II)-based metal affinity chromatography for the isolation of the anticancer agent bleomycin from Streptomyces verticillus culture. J Inorg Biochem 115:198–203
Gu J, Codd R (2015) The resolution of two clinical agents, bleomycin and desferrioxamine B, from a Streptomyces verticillus fermentation mixture using multidimensional immobilised metal ion affinity chromatography. RSC Adv 5:3443
Arthur LB, Peter CF, Steven DB (2001) In vitro activities of daptomycin against 2,789 clinical isolates from 11 North American medical centers. Antimicrob gents Chemother 45:1919–1922
Ng IS, Chimimg Ye, Zhang Z, Lu Y, Jing K (2014) Daptomycin antibiotic production processes in fed-batch fermentation by Streptomyces roseosporus NRRL11379 with precursor effect and medium optimization. Bioprocess Biosyst Eng 37:415–423
Konz D, Klens A, Schörgendorfer K, Marahiel MA (1997) The bacitracin biosynthesis operon of Bacillus licheniformis ATCC 10716: molecular characterization of three multi-modular peptide synthetases. Chem Biol 4:927–937
Murphy T, Roy I, Harrop A, Dixon K, Keshavarz T (2007) Effect of oligosaccharide elicitors on bacitracin A production and evidence of transcriptional level control. J Biotechnol 131:397–403
Shaheen M, Li J, Ross AC, Vederas JC, Jensen SE (2011) Paenibacillus polymyxa PKB1 Produces Variants of polymyxin B-Type antibiotics. Chem Biol 18:1640–1648
Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL (2006) Colistin: the re-emerging antibiotic for multidrug-resistant gram-negative bacterial infections. Lancet Infect Dis 6:589–601
Michalopoulos A, Falagas ME (2008) Colistin and polymyxin B in critical care. Crit Care Clin 24:377–391
Rabanal F, Grau-Campistany A, Vila-Farrés X, Gonzalez-Linares J, Borràs M, Vila J, Manresa A, Cajal Y (2015) A bioinspired peptide scaffold with high antibiotic activity and low in vivo toxicity. Scientific Rep 5:10558
Roberts KD, Azad MAK, Wang J, Horne AS, Thomposon PE, Nation RL, Velkov T, Li J (2016) Antimicrobial activity and toxicity of the major lipopeptide components of polymyxin B and colistin: last-line antibiotics. ACS Infect Dis 1:568–575
Schneditz G, Rentner J, Roier S, Pletz J, Herzog KA, Bücker R, Troeger H, Schild S, Weber H, Breinbauer R, Gorkiewicz G, Högenauer C, Zechner EL (2014) Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. Proc Natl Acad Sci USA 111(36):13181–13186
Jiang J, Gao L, Bie X, Lu Z, Liu H, Zhang C, Lu F, Zhao H (2016) Identification of novel surfactin derivatives from NRPS modification of Bacillus subtilis and its antifungal activity against Fusarium moniliforme. BMC Microbiol 16:31
Robbel L, Marahiel MA (2010) Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J Biol Chem 285(36):27501–27508
Cochrane SA, Vederas JC (2016) Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med Res Rev 36(1):4–31. doi:10.1002/med.21321
Rodrigues L, Banat IM, Teixeira J, Oliveira R (2006) Biosurfactants: potential applications in medicine. J Antimicrob Chemother 57:609–618
Cameotra SS, Makkar RS (2004) Recent applications of biosurfactants as biological and immunological molecules. Curr Opin Microbiol 7:262–266
Reis RS, Pacheco GJ, Pereira AG and Freire DMG (2013) Biosurfactants: production and applications. In: Chamy R (ed) Biodegradation—life of science. InTech
Campos JM, Stamford TLM, Sarubbo LA, Luna JM, Rufino RD, Banat IM (2013) Microbial biosurfactants as additives for food industries. Biotechnol Prog 29:1097–1108
Mukherjee S, Das P, Sen R (2006) Towards commercial production of microbial surfactants. Trends Biotechnol 24(11):509–515
Makkar RS, Cameotra SS, Banat IM (2011) Advances in utilization of renewable substrates for biosurfactant production. AMB Express. 1:5
Van Bogaert INA, Saerens K, De Muynck C, Develter D, Soetaert W, Vandamme EJ (2007) Microbial production and application of sophorolipids. Appl Microbiol Biotechnol 76(1):23–34
Van Bogaert INA, Soetaert W (2011) Sophorolipids. In: Soberón-Chávez G (ed) Biosurfactants: From genes to applications. Springer, Heidelberg
Müller MM, Kügler JH, Henkel M, Gerlitzki M, Hörmann B, Pöhnlein M, Syldatk C, Hausmann R (2012) Rhamnolipids–next generation surfactants? J Biotechnol 162(4):366–380
Marchant R, Banat IM (2012) Microbial biosurfactants: challenges and opportunities for future exploitation. Trends Biotechnol 30(11):558–565
Schwarzer D, Finking R, Marahiel MA (2003) Nonribosomal peptides: from genes to products. Nat Prod Rep 20:275–287
Authors' contributions
MAMN conceived the idea and analyzed the data. MAMN, VELL wrote the paper. Both authors read and approved the final manuscript.
Acknowledgements
We would like to thank to Zuemy Rodríguez Escamilla for their comments and suggestions to this manuscript.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Martínez-Núñez, M.A., López, V.E.L.y. Nonribosomal peptides synthetases and their applications in industry. Sustain Chem Process 4, 13 (2016). https://doi.org/10.1186/s40508-016-0057-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40508-016-0057-6