Abstract
Frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) is a neurodegenerative disease primarily affecting the frontal and/or temporal cortices. However, a growing body of evidence suggests that the cerebellum contributes to biochemical, cognitive, and behavioral changes in FTLD-TDP. To evaluate cerebellar TDP-43 expression and function in FTLD-TDP, we analyzed TDP-43 protein levels and the splicing of a TDP-43 target, STMN2, in the cerebellum of 95 FTLD-TDP cases and 25 non-neurological disease controls. Soluble TDP-43 was decreased in the cerebellum of FTLD-TDP cases but a concomitant increase in insoluble TDP-43 was not seen. Truncated STMN2 transcripts, an indicator of TDP-43 dysfunction, were elevated in the cerebellum of FTLD-TDP cases and inversely associated with TDP-43 levels. Additionally, lower cerebellar TDP-43 associated with a younger age at disease onset. We provide evidence of TDP-43 loss of function in the cerebellum in FTLD-TDP, supporting further investigation into this understudied brain region.
Similar content being viewed by others
Introduction
Frontotemporal dementia (FTD) is an umbrella term for syndromes presenting with deficits in behavior, executive function, and language [1, 2]. Approximately 50% of cases with frontotemporal lobar degeneration (FTLD), the neuropathological diagnosis of FTD, are characterized by cytoplasmic inclusions containing TAR DNA binding protein 43 (TDP-43) in neurons and glia, with a concomitant loss of nuclear TDP-43 [3, 4]. Consequently, TDP-43 loss of function (due to its nuclear depletion) and/or toxic gains of function caused by aggregated cytoplasmic TDP-43 are believed to underlie the neuronal susceptibility and degeneration observed in the frontal and temporal cortices in FTLD with TDP-43 pathology (FTLD-TDP) [5].
The cerebellum has historically been underappreciated in FTLD-TDP given the absence of TDP-43 inclusions and significant neurodegeneration in this neuroanatomical region. However, functional imaging studies show cerebellar involvement in cognitive abilities such as working memory, emotion, language, and attention processing [6, 7]. In addition, extensive connections exist between the cerebellum and cerebrum, including the frontal and temporal lobes [8,9,10]. The discovery of a hexanucleotide repeat expansion in the chromosome 9 open reading frame 72 (C9orf72) gene as the most common genetic cause of FTD and amyotrophic lateral sclerosis (ALS)—often referred to as c9FTD/ALS—stoked interest in the cerebellum. The cerebellum of c9FTD/ALS cases is marked by robust dipeptide-repeat protein pathology and foci of repeat RNA transcripts [11,12,13,14]. Reports that a smaller repeat expansion size in the cerebellum offers a survival advantage [15, 16], and that cerebellar poly(GP) dipeptide repeat protein levels associate with cognitive impairment [14] further support cerebellar involvement in c9FTD/ALS. Moreover, studies uncovered cerebellar transcriptome alterations [17, 18], and progressive atrophy in cerebellar subregions associated with cognitive and motor symptoms in FTLD-TDP cases with or without a C9orf72 repeat expansion [19,20,21,22,23,24]. In aggregate, these data implicate cerebellar anomalies in FTLD-TDP, beyond direct C9orf72-associated pathology, leaving the causes of cerebellar dysfunction in FTLD-TDP unanswered.
Previous studies discovered that TDP-43 suppresses the inclusion of cryptic exons in numerous transcripts [25], including STMN2 [26, 27]. We recently demonstrated that a truncated variant of STMN2 (tSTMN2), generated by the aberrant inclusion of a stop-codon containing cryptic exon, is detected in the central nervous system of ALS and FTLD cases with TDP-43 proteinopathy [28]. Intriguingly, in analyses of RNA sequencing datasets, tSTMN2 was also detected in the cerebellum of some ALS and ALS/FTLD cases [28], suggesting a loss of TDP-43 splicing activity in this region.
Given the mounting evidence implicating cerebellar anomalies in FTLD-TDP, we evaluated TDP-43 splicing function and expression in the cerebellum of a cohort of well-characterized FTLD-TDP cases.
Materials and methods
Study design
Post-mortem brain tissue from individuals with neuropathologically confirmed FTLD-TDP and those without neuropathological features were provided by the Mayo Clinic Florida Brain Bank. All participants or their family members gave written informed consent, and all protocols were approved by the Mayo Clinic Institution Review Board and Ethics Committee. Sample size was determined based on the availability of tissue in our brain bank. A description of patient characteristics is provided Table 1.
RNA extraction and NanoString analysis in post-mortem brain tissue
RNA was extracted from postmortem frozen cerebellum tissue using the RNAeasy Plus Mini Kit (Qiagen) per the manufacturer’s instructions. RIN was assessed using an Agilent 2100 bioanalyzer (Agilent Technologies), and only samples with an RNA integrity number (RIN, ≥ 7.0) were used. Levels of the tSTMN2 transcript were determined using 250 ng of RNA using the NanoString PlexSet platform. Data was analyzed using transcript nSolver4.0 software (NanoString Technologies) and normalized to hypoxanthine phosphoribosyltransferase (HPRT1), a housekeeping gene we have previously used to normalized transcript levels [28]. Probe sequences are as follows: tSTMN2 5’-AGAAGACCTTCGAGAGAAAGGTAGAAAATAAGAATTTGGCTCTCTGTGTGAGCATGTGTGCGTGTGTGCGAGAGAGAGAGACAGACAGCCTGC-3’, and HPRT1 (NM_000194.3), 5’- CTATGACTGTAGATTTTATCAGACTGAAGAGCTATTGTAATGACCAGTCAACAGGGGACATAAAAGTAATTGGTGGAGATGATCTCTCAACTTTAACTGG-3’.
Protein extraction and immunoblotting
Radioimmunoprecipitation assay buffer (RIPA)-soluble and urea-soluble protein fractions were obtained from postmortem cerebellar tissue, as previously described [14]. Approximately 50 mg of tissue were homogenized in cold RIPA buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% sodium deoxycholate, 1% Nonidet P-40, 0.1% sodium dodecyl sulfate, protease and phosphatase inhibitors) and then sonicated on ice. The homogenates were centrifuged at 100,000 × g for 30 min at 4 °C and the supernatant was collected. The pellets were resuspended in RIPA buffer and sonicated. The samples were then centrifuged again and extracted using 7 M urea, sonicated, and centrifuged at 100,000 × g for 30 min at room temperature. Protein concentrations of the RIPA and urea-soluble fractions were determined by Bicinchoninic acid assay (BCA) assay or Bradford assay, respectively. RIPA or urea-soluble protein was diluted in 2X Tris–glycine SDS sample buffer (Life Technologies) and reducing agent (5% beta-mercaptoethanol, Sigma Aldrich) and heat-denatured for 5 min at 95 °C. Samples were run by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on 4–20% Tris–Glycine gels (Life Technologies) and transferred to Polyvinylidene (PVDF) membrane (Millipore). Following transfer, membranes were blocked in 5% non-fat dry milk in Tris buffered saline with Triton (TBS-T, 100 mM Tris–HCl pH 7.5, 140 mM NaCl, 0.1% Triton X-100) and incubated overnight at 4 °C with a rabbit polyclonal anti-TDP-43 antibody (1:1500, 12892-1-AP, ProteinTech) followed by a mouse monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (1:30,000, H86504M, Meridian Life Sciences). Membranes were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000; Jackson ImmunoResearch) and proteins of interest were detected by Enhanced chemiluminescence (ECL, PerkinElmer). Quantitative densitometry on soluble and insoluble protein fractions was performed using Image J and TDP-43 in RIPA-soluble protein fractions were normalized to GAPDH expression.
Statistics
All statistical analyses were done using GraphPad Prism 9 (GraphPad Software). For each figure the type of analysis used, and the number of subjects is indicated in the figure and/or legend.
To compare tSTMN2 RNA and TDP-43 protein in the cerebellum between controls and FTLD-TDP cases, all FTLD-TDP cases combined and the three genotypes separately, were analyzed with single-variable (unadjusted) and multivariable linear regression models (adjusted). Multivariable models were adjusted for age at death, sex and RIN for tSTMN2 RNA, and for age at death and sex for TDP-43 protein. Both tSTMN2 RNA and TDP-43 protein were analyzed on the base 2 logarithmic scale due to their skewed distributions. The regression coefficients (β) and 95% confidence intervals (CIs) were estimated and interpreted as the difference in the means, bases on the 2 logarithmic scale, between all FTLD-TDP cases combined or the individual genotypes and the controls (reference group). P values less than 0.0125 were considered statistically significant after adjusting for the four different statistical test that were performed for all FTLD-TDP and the separate FTLD-TDP groups (no mutation, C9orf72 mutation carriers, GRN mutation carriers) vs. controls.
For associations between TDP-43 protein levels and tSTMN2 RNA, age at disease onset, or disease duration after onset were evaluated in FTLD-TDP cases using single-variable and multivariable linear regression models. Both TDP-43 protein levels and tSTMN2 RNA were analyzed using the base 2 logarithmic scale. The multivariable model examining TDP-43 protein levels and tSTMN2 RNA was adjusted for age, sex and genotype. The model evaluating TDP-43 and age at disease onset was adjusted for sex and genotype, and the model assessing associations between TDP-43 protein and disease duration was adjusted for sex, age at onset and genotype. P values less than 0.0167 were considered statistically significant after adjusting for multiple comparisons.
To compare levels of soluble (RIPA-soluble) or insoluble (urea-soluble) TDP-43 protein a Mann–Whitney test was used.
Results
Truncated STMN2 RNA is elevated in the cerebellum of FTLD-TDP cases
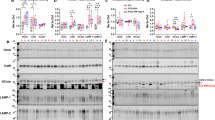
To evaluate cerebellar STMN2 missplicing, we used the NanoString PlexSet platform to measure tSTMN2 RNA in 95 FTLD-TDP cases ([29] with no known FTD-causing mutation, 29 with a C9orf72 repeat expansion and 33 with a mutation in progranulin (GRN)) and in 25 cognitively normal controls pathologically-confirmed to have no TDP-43 pathology (see Table 1). Our control and FTLD-TDP groups were sex-matched, having similar ratios of males to females. However, individuals in our control group were significantly older and had higher RNA integrity numbers (RIN, Table 1). We used a single-variable linear regression model, referred to as the unadjusted analysis, to first determine trends in the unadjusted data and then a multivariable linear regression model, referred to as the adjusted model, to control for possible confounding factors arising from differences between control and FTLD-TDP groups. In unadjusted analysis [β:0.9352, 95% confidence interval (CI): 0.5235 to 1.347, P < 0.0001] and in analysis adjusted for age, sex and RIN (β:0.7475, 95% CI: 0.3797 to 1.115, P = 0.0001), cerebellar tSTMN2 RNA was significantly higher in all FTLD-TDP cases combined or in each separate FTLD-TDP group when compared to controls (Fig. 1A, Table 2).
Truncated STMN2 RNA is elevated, and soluble TDP-43 protein is decreased in the cerebellum of FTLD-TDP cases. A, B tSTMN2 RNA and TDP-43 protein levels were measured in FTLD-TDP cases (n = 95) and non-neurological disease controls (n = 25). A tSTMN2 RNA levels, as measured by NanoString PlexSet platform, are significantly elevated in FTLD-TDP cases. B Soluble TDP-43 protein levels, as quantified from Western blot, are significantly decreased in FTLD-TDP cases. Horizontal bars represent mean 95% confidence intervals (95% CI). P-values and 95% CI from unadjusted linear regression models are shown. Note values for linear regression models adjusted for age, sex and RIN are shown in Table 2. C Associations of TDP-43 protein levels with tSTMN2 RNA levels were examined in FTLD-TDP cases. Regression coefficient (β), 95% CI and P-values from unadjusted linear regression models are shown. Values for linear regression models adjusted for age, sex and genotype are shown in Table 3. D, E TDP-43 protein levels in the cerebellum of FTLD-TDP cases (n = 8) and controls (n = 10) were measured by western blot in the RIPA-soluble (D) and RIPA-insoluble (E) fractions. A representative western-blot (above) and densiometric quantification (below) are presented. Data are presented as mean + / − SEM. P values shown resulting from Mann Whitney test
TDP-43 protein is reduced in the cerebellum in FTLD-TDP cases and associates with elevated truncated STMN2 RNA levels
To test the hypothesis that the presence of tSTMN2 RNA in the cerebellum was caused by suboptimal TDP-43 levels, we examined detergent-soluble TDP-43 protein levels in the cerebellum by Western blot. Compared to controls, a significant or nominally significant decrease in cerebellar TDP-43 was seen in FTLD-TDP cases in unadjusted analysis (β: − 0.4087, 95% CI: − 0.6325 to − 0.1849, P = 0.0004, Fig. 1B, Table 2) and analysis adjusted for age and sex (β: − 0.2701, 95% CI: − 0.4850 to − 0.05514, P = 0.0142, Table 2), respectively. This decrease in soluble TDP-43 appeared to be largely driven by C9orf72 and GRN mutation carriers (Fig. 1B, Table 2). Further confirming the relationship between cerebellar TDP-43 protein and tSTMN2 RNA in FTLD-TDP cases, we found that lower soluble TDP-43 significantly associated with higher tSTMN2 in both unadjusted analysis (β: − 0.2604, 95% CI: − 0.3621 to − 0.1588, P < 0.0001, Fig. 1C, Table 3) and analysis adjusted for age, sex and RIN (β: − 0.2078, 95% CI: − 0.3136 to − 0.1021, P = 0.0002, Table 3).
To determine whether the decrease in soluble TDP-43 in FTLD-TDP cases was caused by its redistribution to the detergent-insoluble (RIPA-insoluble, urea-soluble) fraction, we measured cerebellar TDP-43 in soluble and insoluble fractions for a subset of FTLD-TDP cases and controls. More specifically, we selected the FTLD-TDP cases showing the most drastic depletion of soluble TDP-43 (Fig. 1D) as they would likely best reveal any appreciable shift of TDP-43 from the soluble to the insoluble fraction. As expected, based on our sample selection, soluble TDP-43 was significantly lower in FTLD-TDP cases than in controls; however, no concomitant rise in insoluble TDP-43 was observed (Fig. 1D, E). The lower TDP-43 levels suggest that overall cerebellar TDP-43 expression is decreased in FTLD-TDP.
Lower levels of cerebellar TDP-43 associate with an earlier age at disease onset
To evaluate the clinical significance of cerebellar TDP-43 depletion and elevated tSTMN2 RNA production, we assessed whether detergent soluble cerebellar TDP-43 or tSTMN2 RNA levels associate with age of disease onset or disease duration in FTLD-TDP cases. Lower cerebellar TDP-43 associated with a younger age at disease onset in both unadjusted analysis (β: 0.02175, 95% CI: 0.009906 to 0.03359, P = 0.0005, Fig. 2A, Table 3), and analysis adjusted for sex and genotype (β: 0.01624. 95% CI: 0.003352 to 0.02912, P = 0.0142, Table 3). Although TDP-43 protein levels did not associate with disease duration in unadjusted analysis (β: 0.02088, 95% CI: − 0.003103 to 0.04487, P = 0.0871, Fig. 2B, Table 3), a trend of lower TDP-43 and shorter disease duration was observed in analysis correcting for age, sex and genotype (β: 0.02693, 95% CI: 0.004536 to 0.04932, P = 0.0190, Table 3). Higher levels of cerebellar tSTMN2 RNA significantly associated with an earlier age of disease in unadjusted analysis (β: − 0.02638, 95% CI: − 0.04788 to − 0.004882, P = 0.0168, Additional file 1: Table S1) and trended toward significance in analysis adjusted for RIN and sex (β: − 0.02056, 95% CI: − 0.04029 to − 0.0008248, P = 0.0414, Additional file 1: Table S1). Levels of tSTMN2 RNA did not associate with disease duration in either unadjusted (β: − 0.01976, 95% CI: − 0.06213 to 0.02260, P = 0.3562, Additional file 1: Table S1) or analysis adjusted for age, sex, and RIN (β: − 0.01474, 95% CI: − 0.05858 to 0.02910, P = 0.5052, Additional file 1: Table S1). In aggregate, our data suggest that TDP-43-mediated dysfunction in the cerebellum contributes to FTLD-TDP pathogenesis.
Lower levels of cerebellar TDP-43 associated with an earlier age at disease onset. Associations of TDP-43 protein levels with age at disease onset or disease duration (i.e., time from disease onset to death) were examined in FTLD-TDP cases. A Lower TDP-43 protein significantly associated with younger age at disease onset. B A non-significant trend of lower TDP-43 protein levels and shorter disease duration was observed. Regression coefficients (β), 95% confidence intervals (95% CI) and P-values from unadjusted linear regression models are shown. Values for linear regression models adjusted for age, sex and genotype are shown in Table 3
Discussion
We found decreased TDP-43 protein levels, increased tSTMN2 RNA levels, and a significant inverse association between them in the cerebellum of FTLD-TDP cases. To our knowledge, this is the first report of TDP-43 loss of function in the cerebellum, highlighting that the consequences of a loss of TDP-43 splicing activity may be more extensive than previously thought. Several studies have found dysregulation of gene expression and splicing in the cerebellum of FTLD/ALS cases [15,16,17,18, 30, 31], with one study reporting extensive overlap between gene expression changes in the temporal and frontal cortices and the cerebellum [31]. Similarly, in Alzheimer’s disease and progressive supranuclear palsy, transcriptomic changes detected in the temporal cortex were preserved in the cerebellum [32]. Together, these studies suggest a common mechanism may account for the shared transcriptomic alterations found in distinct brain regions for a given disease. Our data suggest loss of TDP-43 protein levels, and therefore TDP-43 function, may drive these transcriptomic changes in FTLD-TDP. In support of cerebellar TDP-43 dysfunction, 70 genes with differential transcript usage, including polyadenylation, promoter usage and splicing were also identified as targets of TDP-43 [31]. Similarly, TDP-43 was recently found to repress the inclusion of a cryptic exon harbored in the UNC13A gene [33, 34]. The UNC13A transcript with cryptic exon was detected in the frontal and temporal cortices of FTLD/ALS cases with TDP-43 pathology and was also found sparingly in the cerebellum [33, 34].
That we find splicing defects in the cerebellum, a region without significant TDP-43 inclusions, suggest that TDP-43 loss of function is uncoupled from pathology. Reports documenting nuclear TDP-43 clearance in the absence of overt TDP-43 inclusions in regions with TDP-43 pathology thus bear mentioning [29, 35,36,37,38]. It is also noteworthy that TDP-43 nuclear depletion, accompanied by brain atrophy and C9orf72-related pathology, were observed in the temporal lobe of a C9orf72 repeat expansion carrier following surgery for epilepsy [38]. Subsequent post-mortem analysis years later, following the onset of FTD symptoms, revealed TDP-43 inclusions in this individual, suggesting that loss of nuclear TDP-43 is an early pathological event [38]. Further, neurons depleted of TDP-43 in the absence of TDP-43 inclusions were reported to have degenerate morphologies, suggesting that TDP-43 loss of function alone can contribute to neurodegeneration [37, 38]. TDP-43-regulated cryptic exon splice variants were found to accumulate in the hippocampi of Alzheimer’s disease brains containing TDP-43-depleted neurons without TDP-43 inclusions [29]. Likewise, tSTMN2, the UNC13A variant with the cryptic exon and other alternatively spliced genes were detected in TDP-43 negative nuclei isolated from FTD/ALS cases [34, 39]. Taken together, these reports, combined with our present cerebellar data, underscore that TDP-43 nuclear clearance and loss of function can occur independently of inclusion formation and may be an early event in TDP-43 proteinopathy.
We noted that loss of soluble TDP-43 protein was more pronounced in the cerebellum of mutation carriers with the decrease in non-mutation carriers failing to reach statistical significance. However, each FTLD-TDP group (i.e., non-mutation carriers, C9orf72 expansion carriers or GRN mutation carriers) showed a significant accumulation of tSTMN2 RNA arguing that even a minor change in TDP-43 levels results in aberrant STMN2 splicing. These data suggest that tSTMN2 and other cryptic exon-containing transcripts normally suppressed by TDP-43 are sensitive markers of TDP-43 loss of function, thus providing a rationale to study their possible utility as biomarkers for distinguishing patients with FTD caused by TDP-43 proteinopathy vs. tauopathy—an endeavor of great importance to the FTD field.
We additionally observed that lower levels of cerebellar TDP-43 associated with an earlier age at disease onset, suggesting that maintaining cerebellar TDP-43 levels may be protective in FTLD-TDP. We also noted a trend of higher tSTMN2 RNA associating with lower age at disease onset, in agreement with our earlier study finding a significant association between tSTMN2 in the frontal cortex and disease onset in a larger FTLD-TDP cohort [28]. In combination with previously reported transcriptomic and splicing changes [17, 18, 31], and imaging studies demonstrating gross cerebellar atrophy [19,20,21,22,23,24], these findings strongly support a cerebellar contribution to FTLD-TDP pathogenesis.
Conclusions
Overall, we conclude that TDP-43 loss of function may be more pervasive than initially appreciated, and that further study of the role of the cerebellum in the pathogenesis of FTLD-TDP is warranted.
Availability of data and materials
The data used in this study are available from the corresponding authors upon request.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- BCA:
-
Bicinchoninic acid assay
- C9orf72:
-
Chromosome 9 open reading frame 72
- c9FTD/ALS:
-
C9orf72-mediated FTD/ALS
- CI:
-
Confidence interval
- ECL:
-
Enhanced chemiluminescence
- FTD:
-
Frontotemporal dementia
- FTLD:
-
Frontotemporal lobar degeneration
- FTLD-TDP:
-
FTLD with TDP-43 pathology
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- GRN:
-
Progranulin
- HRP:
-
Horseradish peroxidase
- HPRT1:
-
Hypoxanthine Phosphoribosyltransferase
- PVDF:
-
Polyvinylidene
- RIN:
-
RNA integrity number
- RIPA:
-
Radioimmunoprecipitation assay buffer
- SDS-PAGE:
-
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- STMN2:
-
Stathmin-2
- TDP-43:
-
TAR DNA binding protein 43
- TBS-T:
-
Tris buffered saline with Triton
- tSTMN2:
-
Truncated STMN2
- UNC13A:
-
Unc-13 Homolog A
References
Al-Chalabi A, Hardiman O (2013) The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol 9:617–628. https://doi.org/10.1038/nrneurol.2013.203
Neary D, Snowden J, Mann D (2005) Frontotemporal dementia. Lancet Neurol 4:771–780. https://doi.org/10.1016/s1474-4422(05)70223-4
Long JM, Holtzman DM (2019) Alzheimer disease: an update on pathobiology and treatment strategies. Cell 179:312–339. https://doi.org/10.1016/j.cell.2019.09.001
Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM et al (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314:130–133. https://doi.org/10.1126/science.1134108
Tziortzouda P, Van Den Bosch L, Hirth F (2021) Triad of TDP43 control in neurodegeneration: autoregulation, localization and aggregation. Nat Rev Neurosci 22:197–208. https://doi.org/10.1038/s41583-021-00431-1
Allen G, Buxton RB, Wong EC, Courchesne E (1997) Attentional activation of the cerebellum independent of motor involvement. Science 275:1940–1943. https://doi.org/10.1126/science.275.5308.1940
King M, Hernandez-Castillo CR, Poldrack RA, Ivry RB, Diedrichsen J (2019) Functional boundaries in the human cerebellum revealed by a multi-domain task battery. Nat Neurosci 22:1371–1378. https://doi.org/10.1038/s41593-019-0436-x
Buckner RL (2013) The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron 80:807–815. https://doi.org/10.1016/j.neuron.2013.10.044
Habas C, Kamdar N, Nguyen D, Prater K, Beckmann CF, Menon V, Greicius MD (2009) Distinct cerebellar contributions to intrinsic connectivity networks. J Neurosci 29:8586–8594. https://doi.org/10.1523/jneurosci.1868-09.2009
Keser Z, Hasan KM, Mwangi BI, Kamali A, Ucisik-Keser FE, Riascos RF, Yozbatiran N, Francisco GE, Narayana PA (2015) Diffusion tensor imaging of the human cerebellar pathways and their interplay with cerebral macrostructure. Front Neuroanat 9:41. https://doi.org/10.3389/fnana.2015.00041
Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R et al (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77:639–646. https://doi.org/10.1016/j.neuron.2013.02.004
DeJesus-Hernandez M, Finch NA, Wang X, Gendron TF, Bieniek KF, Heckman MG, Vasilevich A, Murray ME, Rousseau L, Weesner R et al (2017) In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers. Acta Neuropathol 134:255–269. https://doi.org/10.1007/s00401-017-1725-7
Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J et al (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126:829–844. https://doi.org/10.1007/s00401-013-1192-8
Gendron TF, van Blitterswijk M, Bieniek KF, Daughrity LM, Jiang J, Rush BK, Pedraza O, Lucas JA, Murray ME, Desaro P et al (2015) Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol 130:559–573. https://doi.org/10.1007/s00401-015-1474-4
DeJesus-Hernandez M, Aleff RA, Jackson JL, Finch NA, Baker MC, Gendron TF, Murray ME, McLaughlin IJ, Harting JR, Graff-Radford NR et al (2021) Long-read targeted sequencing uncovers clinicopathological associations for C9orf72-linked diseases. Brain 144:1082–1088. https://doi.org/10.1093/brain/awab006
van Blitterswijk M, Gendron TF, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Daughrity LM, Murray ME, Heckman MG, Jiang J et al (2015) Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathol 130:863–876. https://doi.org/10.1007/s00401-015-1480-6
Finch NA, Wang X, Baker MC, Heckman MG, Gendron TF, Bieniek KF, Wuu J, DeJesus-Hernandez M, Brown PH, Chew J et al (2017) Abnormal expression of homeobox genes and transthyretin in C9ORF72 expansion carriers. Neurol Genet 3:e161. https://doi.org/10.1212/nxg.0000000000000161
Prudencio M, Belzil VV, Batra R, Ross CA, Gendron TF, Pregent LJ, Murray ME, Overstreet KK, Piazza-Johnston AE, Desaro P et al (2015) Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci 18:1175–1182. https://doi.org/10.1038/nn.4065
Bede P, Hardiman O (2018) Longitudinal structural changes in ALS: a three time-point imaging study of white and gray matter degeneration. Amyotroph Lateral Scler Frontotemporal Degener 19:232–241. https://doi.org/10.1080/21678421.2017.1407795
Chen Y, Kumfor F, Landin-Romero R, Irish M, Hodges JR, Piguet O (2018) Cerebellar atrophy and its contribution to cognition in frontotemporal dementias. Ann Neurol 84:98–109. https://doi.org/10.1002/ana.25271
Mahoney CJ, Downey LE, Ridgway GR, Beck J, Clegg S, Blair M, Finnegan S, Leung KK, Yeatman T, Golden H et al (2012) Longitudinal neuroimaging and neuropsychological profiles of frontotemporal dementia with C9ORF72 expansions. Alzheimers Res Ther 4:41. https://doi.org/10.1186/alzrt144
Rohrer JD, Nicholas JM, Cash DM, van Swieten J, Dopper E, Jiskoot L, van Minkelen R, Rombouts SA, Cardoso MJ, Clegg S et al (2015) Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal dementia Initiative (GENFI) study: a cross-sectional analysis. Lancet Neurol 14:253–262. https://doi.org/10.1016/s1474-4422(14)70324-2
Tan RH, Devenney E, Dobson-Stone C, Kwok JB, Hodges JR, Kiernan MC, Halliday GM, Hornberger M (2014) Cerebellar integrity in the amyotrophic lateral sclerosis-frontotemporal dementia continuum. PLoS ONE 9:e105632. https://doi.org/10.1371/journal.pone.0105632
Whitwell JL, Weigand SD, Boeve BF, Senjem ML, Gunter JL, DeJesus-Hernandez M, Rutherford NJ, Baker M, Knopman DS, Wszolek ZK et al (2012) Neuroimaging signatures of frontotemporal dementia genetics: C9ORF72, tau, progranulin and sporadics. Brain 135:794–806. https://doi.org/10.1093/brain/aws001
Ling JP, Pletnikova O, Troncoso JC, Wong PC (2015) TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349:650–655. https://doi.org/10.1126/science.aab0983
Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, Burberry A, Steinbaugh MJ, Gamage KK, Kirchner R et al (2019) ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci 22:167–179. https://doi.org/10.1038/s41593-018-0300-4
Melamed Z, Lopez-Erauskin J, Baughn MW, Zhang O, Drenner K, Sun Y, Freyermuth F, McMahon MA, Beccari MS, Artates JW et al (2019) Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat Neurosci 22:180–190. https://doi.org/10.1038/s41593-018-0293-z
Prudencio M, Humphrey J, Pickles S, Brown AL, Hill SE, Kachergus JM, Shi J, Heckman MG, Spiegel MR, Cook C et al (2020) Truncated stathmin-2 is a marker of TDP-43 pathology in frontotemporal dementia. J Clin Investig 130:6080–6092. https://doi.org/10.1172/jci139741
Sun M, Bell W, LaClair KD, Ling JP, Han H, Kageyama Y, Pletnikova O, Troncoso JC, Wong PC, Chen LL (2017) Cryptic exon incorporation occurs in Alzheimer’s brain lacking TDP-43 inclusion but exhibiting nuclear clearance of TDP-43. Acta Neuropathol 133:923–931. https://doi.org/10.1007/s00401-017-1701-2
Abramzon YA, Fratta P, Traynor BJ, Chia R (2020) The overlapping genetics of amyotrophic lateral sclerosis and frontotemporal dementia. Front Neurosci 14:42. https://doi.org/10.3389/fnins.2020.00042
Hasan R, Humphrey J, Bettencourt C, Newcombe J, Lashley T, Fratta P, Raj T (2021) Transcriptomic analysis of frontotemporal lobar degeneration with TDP-43 pathology reveals cellular alterations across multiple brain regions. Acta Neuropathol. https://doi.org/10.1007/s00401-021-02399-9
Wang X, Allen M, İş Ö, Reddy JS, Tutor-New FQ, Castanedes Casey M, Carrasquillo MM, Oatman SR, Min Y, Asmann YW et al (2022) Alzheimer’s disease and progressive supranuclear palsy share similar transcriptomic changes in distinct brain regions. J Clin Invest. https://doi.org/10.1172/jci149904
Brown AL, Wilkins OG, Keuss MJ, Hill SE, Zanovello M, Lee WC, Bampton A, Lee FCY, Masino L, Qi YA et al (2022) TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 603:131–137. https://doi.org/10.1038/s41586-022-04436-3
Ma XR, Prudencio M, Koike Y, Vatsavayai SC, Kim G, Harbinski F, Briner A, Rodriguez CM, Guo C, Akiyama T et al (2022) TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature 603:124–130. https://doi.org/10.1038/s41586-022-04424-7
Braak H, Del Tredici K (2018) Anterior Cingulate Cortex TDP-43 Pathology in Sporadic Amyotrophic Lateral Sclerosis. J Neuropathol Exp Neurol 77:74–83. https://doi.org/10.1093/jnen/nlx104
Braak H, Ludolph AC, Neumann M, Ravits J, Del Tredici K (2017) Pathological TDP-43 changes in Betz cells differ from those in bulbar and spinal α-motoneurons in sporadic amyotrophic lateral sclerosis. Acta Neuropathol 133:79–90. https://doi.org/10.1007/s00401-016-1633-2
Nana AL, Sidhu M, Gaus SE, Hwang JL, Li L, Park Y, Kim EJ, Pasquini L, Allen IE, Rankin KP et al (2019) Neurons selectively targeted in frontotemporal dementia reveal early stage TDP-43 pathobiology. Acta Neuropathol 137:27–46. https://doi.org/10.1007/s00401-018-1942-8
Vatsavayai SC, Yoon SJ, Gardner RC, Gendron TF, Vargas JN, Trujillo A, Pribadi M, Phillips JJ, Gaus SE, Hixson JD et al (2016) Timing and significance of pathological features in C9orf72 expansion-associated frontotemporal dementia. Brain 139:3202–3216. https://doi.org/10.1093/brain/aww250
Liu EY, Russ J, Cali CP, Phan JM, Amlie-Wolf A, Lee EB (2019) Loss of nuclear TDP-43 Is associated with decondensation of LINE retrotransposons. Cell Rep 27:1409-1421.e1406. https://doi.org/10.1016/j.celrep.2019.04.003
Funding
This work was supported by NIH grants R35NS097273 (LP), U54NS123743 (LP, MP), P01NS084974 (LP), RF1NS120992 (MP, KAJ), R01AG37491 (KAJ), R01AG063780 (CNC), and the Robert Packard Center for ALS Research at Johns Hopkins (L.P.). YK is supported by Milton Safenowitz Postdoctoral Fellowship Program from the Amyotrophic Lateral Sclerosis Association (21-PDF-582). SP is supported by a BrightFocus ADR Grant (A2020279F). ZKW is supported by a gift from the Donald G. and Jodi P. Herringa Family.
Author information
Authors and Affiliations
Contributions
SP, TFG, LP, CNC, and MP designed the study. YK, YS, and MY performed RNA and protein extractions from human tissue. MP, JMK, JS, and EAT performed NanoString assays. MY, CNC, and SP performed Western blots. MD, BO, NRG-R, BFB, RCP, ZKW, KAJ, and DWD provided human tissue samples as well as pathological, genetic, and clinical information. SP, TFG, CNC, and MP analyzed the data. TFG, and MP performed statistical analysis. SP, TFG, LP, CNC, and MP wrote the manuscript. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All participants or their family members gave written informed consent, and all protocols were approved by the Mayo Clinic Institution Review Board and Ethics Committee.
Consent for publication
Not applicable.
Competing interests
MP serves as a consultant for Target ALS. LP serves as a consultant for Expansion Therapeutics. LP has licensing agreements for frontotemporal dementia mouse models and antibodies. ZKW is partially supported by the NIH/NIA and NIH/NINDS (1U19AG063911, FAIN: U19AG063911), Mayo Clinic Center for Regenerative Medicine, a gift from the Donald G. and Jodi P. Heeringa Family, the Haworth Family Professorship in Neurodegenerative Diseases fund, and The Albertson Parkinson's Research Foundation. ZKW serves as PI or Co-PI on Biohaven Pharmaceuticals, Inc. (BHV4157-206 and BHV3241-301), Neuraly, Inc. (NLY01-PD-1), and Vigil Neuroscience, Inc. (VGL101-01.001 and VGL101-01.002) grants. ZKW serves as Co-PI of the Mayo Clinic APDA Center for Advanced Research and as an external advisory board member for the Vigil Neuroscience, Inc. BO serves as a consultant for Columbia University/Tsumura Inc, MediciNova, and Mitsubishi and have research grants from Columbia University/Tsumura Inc, Biogen, MediciNova, Cytokinetics, Mitsubishi, Calico, and Target ALS.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Associations of cerebellar tSTMN2 RNA with age at onset, and disease duration in FTLD-TDP cases.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Pickles, S., Gendron, T.F., Koike, Y. et al. Evidence of cerebellar TDP-43 loss of function in FTLD-TDP. acta neuropathol commun 10, 107 (2022). https://doi.org/10.1186/s40478-022-01408-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40478-022-01408-6