Abstract
Long non-coding RNAs (lncRNAs) represent an important class of RNAs comprising more than 200 nucleotides, which are produced by RNA polymerase II. Although lacking an open reading framework and protein-encoding activity, lncRNAs can mediate endogenous gene expression by serving as chromatin remodeler, transcriptional or post-transcriptional modulator, and splicing regulator during gene modification. In recent years, increasing evidence shows the significance of lncRNAs in many malignancies, with vital roles in tumorigenesis and cancer progression. Moreover, lncRNAs were also considered potential diagnostic and prognostic markers in cancer. The lncRNA small nuclear RNA host gene 16 (SNHG16), found on chromosome 17q25.1, represents a novel tumor-associated lncRNA. SNHG16 was recently found to exhibit dysregulated expression in a variety of malignancies. There are growing evidence of SNHG16’s involvement in characteristics of cancer, including proliferation, apoptosis, together with its involvement in chemoresistance. In addition, SNHG16 has been described as a promising diagnostic and prognostic biomarker in cancer patients. The current review briefly summarizes recently reported findings about SNHG16 and discuss its expression, roles, mechanisms, and diagnostic and prognostic values in human cancers.
Similar content being viewed by others
Introduction
Cancer remains an important cause of death worldwide, with increasing incidence and mortality [1, 2]. There are approximately 18.1 million new cancer cases and 9.6 million cancer-related deaths worldwide in 2018 [3]. In recent years, although the understanding of molecular mechanisms underlying cancer has increased substantially [4,5,6,7], and treatments for cancer have greatly developed [8, 9], the recurrence and mortality rate of cancer patients are still pessimistic [1]. Therefore, it is imperative to find novel and effective biomarkers and therapeutic targets.
Several recent reports have shown that non-coding RNAs (ncRNAs) contribute to tumorigenesis [10]. NcRNAs are generally divided into two categories by length, including small ncRNAs with length below 200 nucleotides (nts) and long ncRNAs (lncRNAs), which have more than 200 nts [11, 12]. MicroRNAs (miRNAs), a kind of small ncRNAs with 20 to 25 nucleotides, have been shown to negatively regulate the expression of particular key genes and participate in various aspects of cell biology [13, 14]. LncRNAs have attracted increasing attention in recent years. Although lncRNAs represent transcripts without protein-coding potential, they could control gene expression at multiple levels with epigenetic (e.g., DNA methylation, histone modifications, and chromatin remodeling), transcriptional (e.g., recruitment of RNA polymerase II, transcription factors and cofactors), and post-transcriptional (e.g., sponging of miRNAs, alternative splicing, regulating mRNA stability, regulating translation, interacting with proteins) regulations [15,16,17]. Increasing evidence suggests that many lncRNAs show abnormal expression levels in various tumors [18]. LncRNA dysregulation is generally involved in pertinent tumor cellular events, including growth, programmed cell death, metastasis, and stemness [19].
The lncRNA small nuclear RNA host gene 16 (SNHG16) (also named as non-coding RNA expressed in aggressive neuroblastoma [ncRAN]), which contains 2435 nts, is located on chromosome 17q25.1 [20]. There are two splicing variants of SNHG16, including the long form Nbla10727 (with length of 2186 nts) and the short form Nbla12061 (with length of 2087 nts) [21]. SNHG16 was firstly described as a potent oncogenic factor causing poor patient outcome in neuroblastoma [20]. The latter study also showed that elevated SNHG16 amounts contribute to the amplification of the v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) (MYCN) (2p24) locus in this malignancy [20]. Subsequent studies showed that SNHG16 was deregulated and function in several cancers. This review summarizes recently reported findings about SNHG16’s roles in malignancies, including those describing the abnormal expression, molecular functions, and regulatory network of SNHG16, in addition to associated clinical features.
Expression of SNHG16 in cancer
SNHG16 expression is typically increased in a variety cancers, including acute lymphoblastic leukemia, bladder cancer, breast carcinoma, cervical and ovarian cancers, B-cell lymphoma, endometrial carcinoma, esophageal cancer, gastric cancer, glioma, neuroblastoma, lung carcinoma, mouth cancer, osteosarcoma, pancreatic cancer, papillary thyroid cancer, and retinoblastoma (Table 1). Cell lines established from these malignancies also show trends of SNHG16 upregulation. However, in colorectal cancer and hepatocellular carcinoma, there are contrary reports of SNHG16 expression. In hepatocellular carcinoma, the first report by Xu and colleagues [44] demonstrated that SNHG16 amounts are reduced in liver cancer tissue samples and cells. These authors speculated that the site and microenvironment of a given malignancy might provide explanations regarding differences in SNHG16 expression compared with other human cancers. However, after that, at least 5 studies have reported that SNHG16 is upregulated in liver cancer tissue specimens and cells [45, 46]. In colorectal cancer, two reports by Christensen and colleagues [31] and Li and collaborators [30] revealed SNHG16 upregulation in colorectal cancer tissue specimens and cells. On the contrary, another study by Qi et al. [32] reported SNHG16 downregulation in colorectal cancer. Christensen et al. [31] analyzed the above discrepancies in colorectal cancer, and suggested that the methods applied and assessed isoforms are improbable explanations, while RNA quality, tumor cell ratio in tissues, and variations in the used cells could explain these inconsistencies. In the report by Christensen et al., high-quality RNA was tested (median RIN score > 8.9), tumor cells comprised averagely 75% of colorectal cancer tissues, and the HCT116 cell line used was authenticated; these data were not reported by Qi and collaborators.
Regulation of SNHG16 expression in cancer
Several transcription factors can regulate SNHG16 transcription in a positive manner, such as c-Myc, signal Transducer and Activator of Transcription 3 (STAT3), and transcription factor AP-2 alpha (TFAP2A). Li and co-workers [55] showed that SNHG16 is regulated by c-Myc, a transcription factor that exerts its function via recruitment of histone acetyltransferase and induction of RNA polymerase II clearance [64]. Meanwhile, Christensen and colleagues [31] demonstrated that SNHG16 is positively associated with the levels of transcription factors controlled by Wnt in colorectal cancer. β-catenin knockdown results in decreased SNHG16 and c-Myc amounts [31]. In addition, SNHG16 overexpression abrogated the cell function changes caused by c-Myc knockdown. In another study, Christensen et al. [31] found that SNHG16 was positively associated with the expression of the Wnt-regulated transcription factors in colorectal cancer. β-catenin knockdown resulted in decreased expression of SNHG16 and c-Myc [31]. In addition, c-Myc knockdown resulted in SNHG16 downregulation, while its upregulation markedly increased SNHG16 expression [31]. These results strongly indicate that Wnt signaling-controlled transcription factors, including c-Myc, could regulate SNHG16 expression. In addition to c-Myc, STAT3 could also influence SNHG16 expression [31]. Small interfering RNA (si-RNA) knockdown of STAT3 resulted in decreased SNHG16 expression in colorectal cancer cells. Moreover, transcription factor TFAP2A could directly bind with the SNHG16 promotor region and activate its transcription of SNHG16 [34].
Functional activities of SNHG16 in cancer
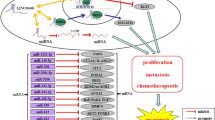
Evidence suggests that several events contribute to malignant features of cells such as sustained growth, resistance to death, induced invasion and metastasis, and increased chemotherapeutic resistance. The hallmarks of cancer are due to mutations producing oncogenes and tumor suppressors. In recent years, SNHG16 has been highlighted with its important functions in mediating these oncogenes and tumor suppressors and generally regulating these cancer characteristics (Table 2).
SNHG16 in cell proliferation
Clearly SNHG16 could regulate the growth and proliferation of various cell lines, and thus regulate the growth of cancers in xenograft models (Table 3). SNHG16 expression is tightly associated with tumor size in several human cancer types. The pan-cyclin-dependent kinases (CDK) inhibitor p21 blocks the cell cycle at G0/G1. SNHG16 knockdown in bladder cancer cells could significantly induce cell cycle arrest at G1 by increasing p21 expression [25]. In agreement, cell cycle arrest at G1 was reported in glioma cells following SNHG16 silencing, with si-SNHG16 exerting its effects via p21 upregulation and cyclin D1 and cyclin B1 downregulation [42].
SNHG16 in cell apoptosis
Apoptosis, autophagy, and necrosis are three major pathways leading to cell death [16, 65]. Multiple reports have demonstrated that SNHG16 inhibits apoptosis in different cancers. These studies demonstrated SNHG16 knockdown induced cell apoptosis by flow cytometry detection. In bladder cancer cells, si-SNHG16 upregulates Bax and caspase-3, while downregulating Bcl-2 [23, 24]. Inhibition of SNHG16 could also induced more cleavage and activation of caspase-3 in glima [41] and oral squamous cell carcinoma cells [55]. In addition, in osteosarcoma [56], pediatric neuroblastoma [52], and colorectal cancer cells [31], downregulation of SNHG16 increased caspase 3/7 activity.
SNHG16 in cancer metastasis
Metastasis represents a common cause of death in most cancers other than the primary tumor. SNHG16 affects cancer metastasis predominantly by controlling epithelial-to-mesenchymal transition (EMT). Knockdown of SNHG16 inhibits EMT in bladder cancer [23], colon cancer [30], esophageal cancer [35], cervical cancer [28], oral squamous cell carcinoma [55], gastric cancer [39], and hepatocellular carcinoma [47], and regulates EMT-associated molecules (upregulates E-cadherin, and downregulates N-cadherin and vimentin).
SNHG16 in chemotherapeutic resistance
Currently, the chemotherapeutic treatment of many cancers is mainly hampered by drug resistance. SNHG16 contributes to chemoresistance in hepatocellular carcinoma. It was shown that siRNA knockdown of SNHG16 reverses sorafenib resistance in Hep3B and HepG2 cell lines [45], and sh-SNHG16 knockdown increases HuH7 and Hep3B cell sensitivities to cisplatin [47], but the underlying mechanisms were not clarified by these two studies. However, another study reported that upregulation of SNHG16 inhibits 5-fluorouracil (5-FU) chemoresistance in Hep3B and HuH7 cells [44], which contrasts the two reports mentioned above. Moreover, tissue and cell SNHG16 levels in the latter report [14] were also different from those reported by several other studies; thus, these findings need further validation.
Mechanisms of SNHG16 action in cancer
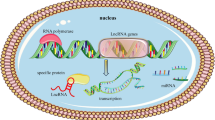
Emerging evidence reveals that the modulatory effects of lncRNAs strongly depend upon their location in cells [66]. LncRNAs located in the cytoplasm may regulate target genes in a post-transcriptional pathway by serving as partners with miRNA [67]. It is widely admitted that SNHG16 is primarily expressed in the cytoplasm of cells [31, 34, 35, 37, 45, 49], and thus acts as a competing endogenous RNA (ceRNA) to affect the expression of key targets. SNHG16 was also reported to be obviously expressed in nucleus [25], and possibly exert its function at the transcriptional level (Fig. 1).
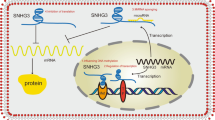
Upstream regulatory and downstream molecular mechanisms underlying SNHG16 in human cancers. a SNHG16 is positively regulated by transcription factors, such as c-Myc, β-catenin, STAT3, and TFAP2A; b SNHG16 binds to EZH2 and recruits EZH2 to p21 promoter, subsequently inhibits p21 expression; c SNHG16 binds to SPI1/PU.1 and recruits SPI1/PU.1 to PARP9 pomoter, subsequently upregulates PARP9 expression; d SNHG16 functions as a ceRNA to regulate multi miRNAs and target genes; e SNHG16 regulates expression of SCD, p-AKT and MMP9, DKK3, and Wnt/β-catenin, with uncovered mechanisms
SNHG16 serves as ceRNA
CeRNA is defined as ncRNAs interact with miRNAs through miRNA response elements (MRE) to inactivate and release the repression of miRNAs to its the target genes. A number of studies demonstrated that SNHG16 functions as a ceRNA to regulate multi miRNAs, including miR-124-3p [22], miR-98 [24, 57], miR-216-5p [28], miR-200a-3p [30], miR-140-5p [35], miR-135a [37], and so on (Table 4). In these studies, luciferase reporter assays and RNA immunoprecipitation (RIP) and/or RNA pull-down assays were carried out to reveal a miRNA-binding site in the SNHG16 sequence, and functional assays suggested that the miRNA and associated target genes control SNHG16’s effects.
SNHG16 interacts with proteins
LncRNAs can interact with proteins to modulate its function. SNHG16 has been reported to directly bind to Enhancer of Zeste Homolog 2 (EZH2) and recruits it to the promoter regions of p21, which subsequently induces histone modification [25]. Another recent study demonstrated that SNHG16 upregulates poly(ADP-ribose) polymerases 9 (PARP9) expression through binding and recruiting transcription factor SPI1/PU.1 to PARP9 pomoter [29].
SNHG16 regulates various genes
In addition, there are some molecules reported to be mediated by SNHG16, including Stearoyl-CoA Desaturase (SCD) [31], p-AKT and matrix metallopeptidase 9 (MMP9) [60], Dickkopf-related protein 3 (DKK3) [39], and Wnt/β-catenin [36], but the underlying regulate mechanisms are not well uncovered. Furthermore, whether SNHG16 has other regulatory functions still needs further investigation.
SNHG16 as a biomarker in cancer
SNHG16 serves as a diagnostic biomarker in cancer
LncRNA detection in body fluids, including serum and plasma specimens, provides a novel avenue for early noninvasive cancer diagnosis. SNHG16 was shown to be relatively stable under freeze-thaw cycles or long-term storage, indicating an advantage for SNHG16 as a potentially effective diagnostic biomarker in malignancies [68]. SNHG16’s diagnostic value was investigated in bladder cancer, with elevated plasma, serum, and serum exosome amounts detected in bladder cancer cases compared with healthy controls. Indeed, SNHG16 was shown to be markedly elevated in plasma specimens from 26 bladder cancer cases vs. 15 control patients [24], as well as in serum samples from 120 bladder cancer cases vs. 52 healthy subjects [68]. However, the diagnostic value of SNHG16 was not evaluated in the two above studies, as areas under the receiver operating characteristic (ROC) curves (AUC values) were not shown. In another study, Zhang and collaborators [69] reported increased SNHG16 amounts in the serum exosomes of 100 breast cancer cases in comparison with 100 healthy controls. The AUC was 0.681, which indicates that the accuracy of breast cancer classification based on SNHG16 is low. To date, there is no evidence indicating that SNHG16 alone might constitute an ideal diagnostic biomarker in cancer.
The diagnostic value of SNHG16 was further investigated in combination with other lncRNAs in bladder cancer. Duan and colleagues [68] obtained an AUC for a three-lncRNA panel (SNHG16, MEG3, and MALAT1) of 0.865 in the training set comprising 240 serum samples (52 healthy individuals, and 68 non-cancer and 120 bladder cancer cases) and 0.828 in the validation set composed of 200 serum samples (48 healthy individuals, and 52 non-cancer and 100 bladder cancer cases). This three-lncRNA combination showed AUC values for Ta, T1, and T2-T4 of 0.778, 0.805, and 0.880, respectively, upon serum detection. In another study, Zhang et al. [69] identified a lncRNA panel in serum exosomes for diagnosing and predicting recurrence in bladder cancer using a training set of 100 serum exosome samples (50 healthy controls and 50 bladder cancer patients) and a validation cohort of 320 serum exosome specimens (160 healthy controls and 160 bladder cancer patients). They reported, in the training cohort, an AUC for a three-lncRNA combination (SNHG16, PCAT-1 and UBC1) of 0.857; the accuracy of this lncRNA panel to differentiate between healthy individuals and bladder cancer patients was 0.815 (sensitivity and specificity of 0.85 and 0.78, respectively). The corresponding AUC values for the lncRNA combination in Ta, T1, and T2-T4 cases were 0.760, 0.827, and 0.878, respectively. In the validation cohort, an AUC for the three-lncRNA combination of 0.826 was obtained, and sensitivity and specificity were 0.80 and 0.75, respectively. Moreover, the three-lncRNA combination had an AUC significantly elevated in comparison with urine cytology (0.574), a biomarker with commendable sensitivity in detecting high- but not low-grade bladder cancers, indicating the potential of the above lncRNA panel as an effective biomarker for bladder cancer diagnosis. Collectively, the above findings suggested that the SNHG16-combined-lncRNA panels could differentiate bladder cancer patients from healthy individuals with great accuracy, which warrants further multicenter trials with larger clinical samples for confirmation. However, SNHG16 is found in multiple cancer types and less likely to help distinguish the specific origins of tumors. Thus, further studies should be performed to analyze the diagnostic value of SNHG16 in body fluids in combination with other specific biomarkers in different cancers.
SNHG16 serves as a prognostic marker in cancer
It was shown that abnormal SNHG16 expression is closely related to cancer prognosis (Table 5). High SNHG16 amounts were shown to be significantly related to poor overall survival bladder cancer, cervical cancer, endometrial carcinoma, esophageal cancer, stomach cancer, glioma, liver cancer, neuroblastoma, lung cancer, osteosarcoma, and ovarian and pancreatic cancers. SNHG16 expression independently predicts prognosis in these cancers. Additionally, SNHG16 upregulation was also shown to be related to poor progression free survival (PFS) in glioma, liver cancer, and lung cancer. However, in colon cancer, it was reported that SNHG16 downregulation is related to shorter overall survival (OS) of patients. Apart from survival data, other clinical features related to SNHG16 expression have been reported. In bladder cancer, high tumor SNHG16 levels are significantly related to tumor stage, histological grade, and lymph node metastasis. In cervical cancer, SNHG16 overexpression is related to tumor size and differentiation, as well as lymph node metastasis and FIGO stage. In esophageal cancer, cases expressing high SNHG16 amounts show elevated tumor stage, clinical stage, and lymph node metastasis. In ovarian cancer, high levels of SNHG16 are linked to clinical stage and distant metastasis. In pancreatic cancer, SNHG16 upregulation is associated with TNM stage, tumor differentiation, and distant metastasis. High SNHG16 levels are also associated with tumor size and TNM stage in glioma and hepatocellular carcinoma. Jointly, the above findings indicate that SNHG16 might represent an independent parameter for predicting prognosis in various cancers, although further large trials are still required for confirmation.
Conclusions
Numerous studies have confirmed that lncRNAs have critical functions in human cancer formation and progression. SNHG16, also named ncRAN, was initially reported as a potent oncogene in neuroblastoma. Since then, SNHG16 upregulation has been demonstrated in major human cancers. However, published SNHG16 expression patterns vary, and studies assessing the role of SNHG16 have reported conflicting results even in the same type of cancer (e.g., colorectal or liver cancer). SNHG16 contributes to the regulation of biological events in cells such as proliferation, apoptosis, malignancy potential, and chemoresistance. Thus, SNHG16 may represent a potential therapeutic candidate in a variety of cancer types. Mechanistically, post-transcriptional regulation of genes via ceRNAs appears to be an important mechanism underlying the effects of SNHG16. SNHG16 is mostly found in the cytosol, and sponges multiple miRNAs (more than 25 miRNAs reported so far) for target gene regulation. However, the overall mechanism underlying SNHG16 dysregulation in malignancies is not yet completely understood. In relation to clinical practice, SNHG16 dysregulation shows associations with clinical phenotypes as well as patient survival in many cancers, suggesting that SNHG16 might be a prognostic marker. SNHG16 might also represent a promising noninvasive diagnostic marker in cancer in combination with other specific biomarkers, because it is found in body fluids, including plasma, serum, and serum exosomes. Taken together, data from previous reports and the The Cancer Genome Atlas (TCGA) database indicate the upregulation of and an oncogenic role for SNHG16 in various cancers, although discrepant findings require clarification. In addition, further research is needed to facilitate the translation of basic science research evaluating SNHG16 into clinical benefits.
Availability of data and materials
Not applicable.
Abbreviations
- 5-FU:
-
5-fluorouracil
- AUC:
-
Areas under the receiver operating characteristic curves
- CDK:
-
Cyclin-dependent kinases
- ceRNA:
-
Competing endogenous RNA
- DKK3:
-
Dickkopf-related protein 3
- EMT:
-
Epithelial-to-mesenchymal transition
- EZH2 :
-
Enhancer of Zeste Homolog 2
- MMP9:
-
Matrix metallopeptidase 9
- miRNAs:
-
MicroRNAs
- MRE:
-
MiRNA response elements
- MYCN:
-
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
- ncRAN:
-
Non-coding RNA expressed in aggressive neuroblastoma
- ncRNAs:
-
Non-coding RNAs
- OS:
-
Overall survival
- PARP 9:
-
Poly(ADP-ribose) polymerases 9
- PFS:
-
Progression free survival
- RIP:
-
RNA immunoprecipitation
- ROC:
-
Receiver operating characteristic
- SCD:
-
Stearoyl-CoA Desaturase
- siRNA:
-
Small interfering RNA
- SNHG16:
-
Small nuclear RNA host gene 16
- STAT3:
-
Signal Transducer and Activator of Transcription
- TCGA:
-
The Cancer Genome Atlas
- TFAP2A:
-
Transcription factor AP-2 alpha
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–85.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Singh S, Chouhan S, Mohammad N, Bhat MK. Resistin causes G1 arrest in colon cancer cells through upregulation of SOCS3. FEBS Lett. 2017;591(10):1371–82.
Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. 2020;38(2):167–97.
Mohammad N, Malvi P, Meena AS, Singh SV, Chaube B, Vannuruswamy G, Kulkarni MJ, Bhat MK. Cholesterol depletion by methyl-beta-cyclodextrin augments tamoxifen induced cell death by enhancing its uptake in melanoma. Mol Cancer. 2014;13:204.
Singh SV, Ajay AK, Mohammad N, Malvi P, Chaube B, Meena AS, Bhat MK. Proteasomal inhibition sensitizes cervical cancer cells to mitomycin C-induced bystander effect: the role of tumor microenvironment. Cell Death Dis. 2015;6:e1934.
Mohammad N, Singh SV, Malvi P, Chaube B, Athavale D, Vanuopadath M, Nair SS, Nair B, Bhat MK. Strategy to enhance efficacy of doxorubicin in solid tumor cells by methyl-beta-cyclodextrin: involvement of p53 and Fas receptor ligand complex. Sci Rep. 2015;5:11853.
Yu JX, Upadhaya S, Tatake R, Barkalow F, Hubbard-Lucey VM. Cancer cell therapies: the clinical trial landscape. [published online ahead of print, 2020 May 26]. Nat Rev Drug Discov. 2020. https://doi.org/10.1038/d41573-020-00099-9.
Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58.
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208.
Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang X, Cui Y. Long non-coding RNA MIAT in development and disease: a new player in an old game. J Biomed Sci. 2018;25(1):23.
Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther. 2014;15(11):1444–55.
Muhammad N, Bhattacharya S, Steele R, Ray RB. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7(36):58595–605.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41.
Su M, Xiao Y, Ma J, Cao D, Zhou Y, Wang H, Liao Q, Wang W. Long non-coding RNAs in esophageal cancer: molecular mechanisms, functions, and potential applications. J Hematol Oncol. 2018;11(1):118.
Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712.
Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer. 2016;15(1):50.
Chen WK, Yu XH, Yang W, Wang C, He WS, Yan YG, Zhang J, Wang WJ. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50(1):e12313.
Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu Y, Ozaki T, Isogai E, Nakamura Y, Koda T, et al. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int J Oncol. 2009;34(4):931–8.
Zhou Z, Zhu Y, Gao G, Zhang Y. Long noncoding RNA SNHG16 targets miR-146a-5p/CCL5 to regulate LPS-induced WI-38 cell apoptosis and inflammation in acute pneumonia. Life Sci. 2019;228:189–97.
Yang T, Jin X, Lan J, Wang W. Long non-coding RNA SNHG16 has tumor suppressing effect in acute lymphoblastic leukemia by inverse interaction on hsa-miR-124-3p. IUBMB Life. 2019;71(1):134–42.
Peng H, Li H. The encouraging role of long noncoding RNA small nuclear RNA host gene 16 in epithelial-mesenchymal transition of bladder cancer via directly acting on miR-17-5p/metalloproteinases 3 axis. Mol Carcinog. 2019;58(8):1465–80.
Feng F, Chen A, Huang J, Xia Q, Chen Y, Jin X. Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/beta-catenin pathway axis. J Cell Biochem. 2018;119(11):9408–18.
Cao X, Xu J, Yue D. LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther. 2018;25(1–2):10–7.
Zhu Y, Yu M, Li Z, Kong C, Bi J, Li J, Gao Z. ncRAN, a newly identified long noncoding RNA, enhances human bladder tumor growth, invasion, and survival. Urology. 2011;77(2):510.e511–5.
Cai C, Huo Q, Wang X, Chen B, Yang Q. SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun. 2017;485(2):272–8.
Zhu H, Zeng Y, Zhou CC, Ye W. SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the tumorigenesis of cervical cancer cells. Arch Biochem Biophys. 2018;637:1–8.
Tao L, Wang X, Zhou Q. Long noncoding RNA SNHG16 promotes the tumorigenicity of cervical cancer cells by recruiting transcriptional factor SPI1 to upregulate PARP9. Cell Biol Int. 2019;44(3):773–84.
Li Y, Lu Y, Chen Y. Long non-coding RNA SNHG16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer via sponging miR-200a-3p. Biosci Rep. 2019;39(5):BSR20182498.
Christensen LL, True K, Hamilton MP, Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen JB, Pedersen JS, et al. SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism. Mol Oncol. 2016;10(8):1266–82.
Qi P, Xu MD, Ni SJ, Shen XH, Wei P, Huang D, Tan C, Sheng WQ, Zhou XY, Du X. Down-regulation of ncRAN, a long non-coding RNA, contributes to colorectal cancer cell migration and invasion and predicts poor overall survival for colorectal cancer patients. Mol Carcinog. 2015;54(9):742–50.
Zhu Q, Li Y, Guo Y, Hu L, Xiao Z, Liu X, Wang J, Xu Q, Tong X. Long non-coding RNA SNHG16 promotes proliferation and inhibits apoptosis of diffuse large B-cell lymphoma cells by targeting miR-497-5p/PIM1 axis. J Cell Mol Med. 2019;23(11):7395–405.
Zhang G, Ma A, Jin Y, Pan G, Wang C. LncRNA SNHG16 induced by TFAP2A modulates glycolysis and proliferation of endometrial carcinoma through miR-490-3p/HK2 axis. Am J Transl Res. 2019;11(11):7137–45.
Zhang K, Chen J, Song H, Chen LB. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget. 2018;9(1):1028–40.
Han GH, Lu KJ, Wang P, Ye J, Ye YY, Huang JX. LncRNA SNHG16 predicts poor prognosis in ESCC and promotes cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018;22(12):3795–803.
Wang X, Kan J, Han J, Zhang W, Bai L, Wu H. LncRNA SNHG16 functions as an oncogene by sponging MiR-135a and promotes JAK2/STAT3 signal pathway in gastric Cancer. J Cancer. 2019;10(4):1013–22.
Lian D, Amin B, Du D, Yan W. Enhanced expression of the long non-coding RNA SNHG16 contributes to gastric cancer progression and metastasis. Cancer Biomark. 2017;21(1):151–60.
Zhou C, Zhao J, Liu J, Wei S, Xia Y, Xia W, Bi Y, Yan Z, Huang H. LncRNA SNHG16 promotes epithelial- mesenchymal transition via down-regulation of DKK3 in gastric cancer. Cancer Biomark. 2019;26(4):393–401.
Pang W, Zhai M, Wang Y, Li Z. Long noncoding RNA SNHG16 silencing inhibits the aggressiveness of gastric cancer via upregulation of microRNA-628-3p and consequent decrease of NRP1. Cancer Manag Res. 2019;11:7263–77.
Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen JJ, Chen WJ, Sun WY, Liu XM, Chen JB. LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in Glioma. Cell Physiol Biochem. 2018;45(5):1975–85.
Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 Exerts Oncogenic Functions in Promoting Proliferation of Glioma Through Suppressing p21. Pathol Oncol Res. 2019;26(2):1021–8.
Zhou XY, Liu H, Ding ZB, Xi HP, Wang GW. lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway. Genomics. 2020;112(1):1021–9.
Xu F, Zha G, Wu Y, Cai W, Ao J. Overexpressing lncRNA SNHG16 inhibited HCC proliferation and chemoresistance by functionally sponging hsa-miR-93. Onco Targets Ther. 2018;11:8855–63.
Guo Z, Zhang J, Fan L, Liu J, Yu H, Li X, Sun G. Long noncoding RNA (lncRNA) small Nucleolar RNA host gene 16 (SNHG16) predicts poor prognosis and Sorafenib resistance in hepatocellular carcinoma. Med Sci Monit. 2019;25:2079–86.
Li W, Xu W, Song JS, Wu T, Wang WX. LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19 axis in hepatocellular carcinoma. Neoplasma. 2019;66(3):397–404.
Li S, Peng F, Ning Y, Jiang P, Peng J, Ding X, Zhang J, Jiang T, Xiang S. SNHG16 as the miRNA let-7b-5p sponge facilitates the G2/M and epithelial-mesenchymal transition by regulating CDC25B and HMGA2 expression in hepatocellular carcinoma. J Cell Biochem. 2020;121(3):2543–58.
Xie X, Xu X, Sun C, Yu Z. Long intergenic noncoding RNA SNHG16 interacts with miR-195 to promote proliferation, invasion and tumorigenesis in hepatocellular carcinoma. Exp Cell Res. 2019;383(1):111501.
Zhong JH, Xiang X, Wang YY, Liu X, Qi LN, Luo CP, Wei WE, You XM, Ma L, Xiang BD, et al. The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. J Cell Physiol. 2020;235(2):1090–102.
Chen H, Li M, Huang P. LncRNA SNHG16 promotes hepatocellular carcinoma proliferation, migration and invasion by regulating miR-186 expression. J Cancer. 2019;10(15):3571–81.
Bao J, Zhang S, Meng Q, Qin T. SNHG16 silencing inhibits neuroblastoma progression by Downregulating HOXA7 via sponging miR-128-3p. Neurochem Res. 2020;45(4):825–36.
Yu Y, Chen F, Yang Y, Jin Y, Shi J, Han S, Chu P, Lu J, Tai J. Wang S, et al: lncRNA SNHG16 is associated with proliferation and poor prognosis of pediatric neuroblastoma. Int J Oncol. 2019;55(1):93–102.
Han W, Du X, Liu M, Wang J, Sun L, Li Y. Increased expression of long non-coding RNA SNHG16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int J Biol Macromol. 2019;121:270–8.
Yu L, Chen D, Song J. LncRNA SNHG16 promotes non-small cell lung cancer development through regulating EphA2 expression by sponging miR-520a-3p. Thorac Cancer. 2020;11(3):603–s.
Li S, Zhang S, Chen J. C-Myc induced upregulation of long non-coding RNA SNHG16 enhances progression and carcinogenesis in oral squamous cell carcinoma. Cancer Gene Ther. 2019;26(11-12):400–10.
Su P, Mu S, Wang Z. Long noncoding RNA SNHG16 promotes osteosarcoma cells migration and invasion via sponging miRNA-340. DNA Cell Biol. 2019;38(2):170–5.
Liao S, Xing S, Ma Y. LncRNA SNHG16 sponges miR-98-5p to regulate cellular processes in osteosarcoma. Cancer Chemother Pharmacol. 2019;83(6):1065–74.
Zhu C, Cheng D, Qiu X, Zhuang M, Liu Z. Long noncoding RNA SNHG16 promotes cell proliferation by sponging MicroRNA-205 and Upregulating ZEB1 expression in osteosarcoma. Cell Physiol Biochem. 2018;51(1):429–40.
Wang X, Hu K, Chao Y, Wang L. LncRNA SNHG16 promotes proliferation, migration and invasion of osteosarcoma cells by targeting miR-1301/BCL9 axis. Biomed Pharmacother. 2019;114:108798.
Yang XS, Wang GX, Luo L. Long non-coding RNA SNHG16 promotes cell growth and metastasis in ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(3):616–22.
Liu S, Zhang W, Liu K, Liu Y. LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p. Biomed Pharmacother. 2019;114:108862.
Wen Q, Zhao L, Wang T, Lv N, Cheng X, Zhang G, Bai L. LncRNA SNHG16 drives proliferation and invasion of papillary thyroid cancer through modulation of miR-497. Onco Targets Ther. 2019;12:699–708.
Xu C, Hu C, Wang Y, Liu S. Long noncoding RNA SNHG16 promotes human retinoblastoma progression via sponging miR-140-5p. Biomed Pharmacother. 2019;117:109153.
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–12.
Chen Q, Kang J, Fu C. The independence of and associations among apoptosis, autophagy, and necrosis. Signal Transduct Target Ther. 2018;3:18.
Schmitt AM, Chang HY. Long noncoding RNAs in Cancer pathways. Cancer Cell. 2016;29(4):452–63.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7.
Duan W, Du L, Jiang X, Wang R, Yan S, Xie Y, Yan K, Wang Q, Wang L, Zhang X, et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7(48):78850–8.
Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J, Yan K, Duan W, Zhao Y, Wang Y, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23(2):1396–405.
Acknowledgements
We thank Dr. Zhiqiang Xiao for the helpful discussion.
Funding
This study was supported by Natural Science Foundation of Hunan Province (Grant No. 2019JJ50968, 2019JJ50358); National Natural Science Foundation of China (Grant No. 81802947); Changsha Science and Technology Board (kq1801110).
Author information
Authors and Affiliations
Contributions
All authors have contributed to the preparation of this manuscript. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there is no conflict of interest regarding the publication of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xiao, Y., Xiao, T., Ou, W. et al. LncRNA SNHG16 as a potential biomarker and therapeutic target in human cancers. Biomark Res 8, 41 (2020). https://doi.org/10.1186/s40364-020-00221-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40364-020-00221-4