Abstract
Background
As has been illustrated that long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) are potential regulators in the occurrence and progression of human cancers. LncRNA SNHG16 has been identified as an oncogene involved in the progression of human cancers. However, neither the function nor the underlying molecular mechanism of SNHG16 in osteosarcoma has been discovered.
Purpose
The aim of the study is to explore the role and molecular regulation mechanism of SNHG16 in osteosarcoma.
Methods
The expression of SNHG16 in HNSCC tissues and cells was detected by RT-qPCR assay. The biological function of SNHG16 in osteosarcoma was measured by CCK-8, cell cycle, cell apoptosis and transwell assays. The interaction between SNHG16 and miR-98-5p was studied by luciferase reporter and RIP assays.
Results
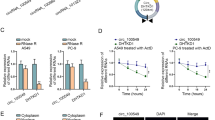
The ectopic expression of SNHG16 was found in osteosarcoma tissues and cell lines, which indicated poor prognosis and lower overall survival rate of osteosarcoma patients. Knockdown of SNHG16 inhibited cell proliferation, migration, invasion, cell cycle and promoted apoptosis in osteosarcoma. It was demonstrated that SNHG16 directly interacts with miR-98-5p. What’s more, we found a significantly negative correlation between SNHG16 and miR-98-5p expression. Finally, rescue experiments revealed that inhibition of miR-98-5p attenuated SNHG16 knockdown-mediated effects on cellular processes in osteosarcoma.
Conclusions
LncRNA SNHG16 regulated cellular processes in osteosarcoma by sponging miR-98-5p, and SNHG16 may be a new and effective molecular therapeutic target for osteosarcoma.




Similar content being viewed by others
References
Benjamin RS (2015) Osteosarcoma: better treatment through better trial design. Lancet Oncol 16(1):12–13. https://doi.org/10.1016/S1470-2045(14)71186-6
Clark JC, Dass CR, Choong PF (2008) A review of clinical and molecular prognostic factors in osteosarcoma. J Cancer Res Clin Oncol 134(3):281–297. https://doi.org/10.1007/s00432-007-0330-x
Durfee RA, Mohammed M, Luu HH (2016) Review of osteosarcoma and current management. Rheumatol Ther 3(2):221–243. https://doi.org/10.1007/s40744-016-0046-y
Ando K, Heymann M-F, Stresing V, Mori K, Rédini F, Heymann D (2013) Current therapeutic strategies and novel approaches in osteosarcoma. Cancers 5(4):591–616. https://doi.org/10.3390/cancers5020591
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136(4):629–641. https://doi.org/10.1016/j.cell.2009.02.006
Ulitsky I (2016) Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet 17(10):601–614. https://doi.org/10.1038/nrg.2016.85
Fatica A, Bozzoni I (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 15(1):7–21. https://doi.org/10.1038/nrg3606
Slaby O, Laga R, Sedlacek O (2017) Therapeutic targeting of non-coding RNAs in cancer. Biochem J 474(24):4219–4251. https://doi.org/10.1042/BCJ20170079
Kondo Y, Shinjo K, Katsushima K (2017) Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci 108(10):1927–1933. https://doi.org/10.1111/cas.13342
DiStefano JK (2018) The emerging role of long noncoding RNAs in human disease. Methods Mol Biol 1706:91–110. https://doi.org/10.1007/978-1-4939-7471-9_6
Chen Y, Du H, Bao L, Liu W (2018) LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med 15(3):238–250. https://doi.org/10.20892/j.issn.2095-3941.2017.0174
Li XL, Subramanian M, Jones MF, Chaudhary R, Singh DK, Zong X, Gryder B, Sindri S, Mo M, Schetter A, Wen X, Parvathaneni S, Kazandjian D, Jenkins LM, Tang W, Elloumi F, Martindale JL, Huarte M, Zhu Y, Robles AI, Frier SM, Rigo F, Cam M, Ambs S, Sharma S, Harris CC, Dasso M, Prasanth KV, Lal A (2017) Long noncoding RNA PURPL suppresses Basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep 20(10):2408–2423. https://doi.org/10.1016/j.celrep.2017.08.041
Ye K, Wang S, Zhang H, Han H, Ma B, Nan W (2017) Long NONCODING RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem 118(12):4772–4781. https://doi.org/10.1002/jcb.26145
Li J, Wu QM, Wang XQ, Zhang CQ (2017) Long noncoding RNA miR210HG sponges miR-503 to facilitate osteosarcoma cell invasion and metastasis. DNA Cell Biol 36(12):1117–1125. https://doi.org/10.1089/dna.2017.3888
Sun L, Yang C, Xu J, Feng Y, Wang L, Cui T (2016) Long noncoding RNA EWSAT1 promotes osteosarcoma cell growth and metastasis through suppression of MEG3 expression. DNA Cell Biol 35(12):812–818. https://doi.org/10.1089/dna.2016.3467
Zhao W, Fu H, Zhang S, Sun S, Liu Y (2018) LncRNA SNHG16 drives proliferation, migration, and invasion of hemangioma endothelial cell through modulation of miR-520d-3p/STAT3 axis. Cancer Med. https://doi.org/10.1002/cam4.1562
Lu YF, Cai XL, Li ZZ, Lv J, Xiang YA, Chen JJ, Chen WJ, Sun WY, Liu XM, Chen JB (2018) LncRNA SNHG16 functions as an oncogene by sponging MiR-4518 and up-regulating PRMT5 expression in glioma. Cell Physiol Biochem 45(5):1975–1985. https://doi.org/10.1159/000487974
Cao X, Xu J, Yue D (2018) LncRNA-SNHG16 predicts poor prognosis and promotes tumor proliferation through epigenetically silencing p21 in bladder cancer. Cancer Gene Ther 25(1–2):10–17. https://doi.org/10.1038/s41417-017-0006-x
Zhu C, Cheng D, Qiu X, Zhuang M, Liu Z (2018) Long noncoding RNA SNHG16 promotes cell proliferation by sponging microRNA-205 and upregulating ZEB1 expression in osteosarcoma. Cell Physiol Biochem 51(1):429–440. https://doi.org/10.1159/000495239
Su P, Mu S, Wang Z (2019) Long noncoding RNA SNHG16 promotes osteosarcoma cells migration and invasion via sponging miRNA-340. DNA Cell Biol 38(2):170–175. https://doi.org/10.1089/dna.2018.4424
Ba Z, Gu L, Hao S, Wang X, Cheng Z, Nie G (2018) Downregulation of lncRNA CASC2 facilitates osteosarcoma growth and invasion through miR-181a. Cell Prolif https://doi.org/10.1111/cpr.12409
Sui CJ, Zhou YM, Shen WF, Dai BH, Lu JJ, Zhang MF, Yang JM (2016) Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J Mol Med (Berl) 94(11):1281–1296. https://doi.org/10.1007/s00109-016-1442-z
Hirata H, Hinoda Y, Shahryari V, Deng G, Nakajima K, Tabatabai ZL, Ishii N, Dahiya R (2015) Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res 75(7):1322–1331. https://doi.org/10.1158/0008-5472.CAN-14-2931
Wu F, Mo Q, Wan X, Dan J, Hu H (2017) NEAT1/has-mir-98-5p/MAPK6 axis is involved in non-small-cell lung cancer (NSCLC) development. J Cell Biochem. https://doi.org/10.1002/jcb.26442
Fu Y, Liu X, Chen Q, Liu T, Lu C, Yu J, Miao Y, Wei J (2018) Downregulated miR-98-5p promotes PDAC proliferation and metastasis by reversely regulating MAP4K4. J Exp Clin Cancer Res 37(1):130. https://doi.org/10.1186/s13046-018-0807-2
Wang Y, Bao W, Liu Y, Wang S, Xu S, Li X, Li Y, Wu S (2018) miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis 9(5):447. https://doi.org/10.1038/s41419-018-0390-7
Ye H, Lin J, Yao X, Li Y, Lin X, Lu H (2018) Overexpression of long non-coding RNA NNT-AS1 correlates with tumor progression and poor prognosis in osteosarcoma. Cell Physiol Biochem 45(5):1904–1914. https://doi.org/10.1159/000487966
Zeng SX, Cai QC, Guo CH, Zhi LQ, Dai X, Zhang DF, Ma W (2017) High expression of TRIM29 (ATDC) contributes to poor prognosis and tumor metastasis by inducing epithelial mesenchymal transition in osteosarcoma. Oncol Rep 38(3):1645–1654. https://doi.org/10.3892/or.2017.5842
D’Angelo B, Benedetti E, Cimini A, Giordano A (2016) MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res 36(11):5571–5575. https://doi.org/10.21873/anticanres.11142
Setijono SR, Park M, Kim G, Kim Y, Cho KW, Song SJ (2018) miR-218 and miR-129 regulate breast cancer progression by targeting Lamins. Biochem Biophys Res Commun 496(3):826–833. https://doi.org/10.1016/j.bbrc.2018.01.146
Zhou W, He L, Dai Y, Zhang Y, Wang J, Liu B (2018) MicroRNA-124 inhibits cell proliferation, invasion and migration by targeting CAV1 in bladder cancer. Exp Ther Med 16(4):2811–2820. https://doi.org/10.3892/etm.2018.6537
Wang Y, Tian Y (2018) miR-206 inhibits cell proliferation, migration, and invasion by targeting BAG3 in human cervical cancer. Oncol Res. https://doi.org/10.3727/096504017X15143731031009
Lu C, Peng K, Guo H, Ren X, Hu S, Cai Y, Han Y, Ma L, Xu P (2018) miR-18a-5p promotes cell invasion and migration of osteosarcoma by directly targeting IRF2. Oncol Lett 16(3):3150–3156. https://doi.org/10.3892/ol.2018.9032
Yang D, Liu G, Wang K (2015) miR-203 acts as a tumor suppressor gene in osteosarcoma by regulating RAB22A. PLoS One 10(9):e0132225. https://doi.org/10.1371/journal.pone.0132225
Feng F, Chen A, Huang J, Xia Q, Chen Y, Jin X (2018) Long noncoding RNA SNHG16 contributes to the development of bladder cancer via regulating miR-98/STAT3/Wnt/beta-catenin pathway axis. J Cell Biochem 119(11):9408–9418. https://doi.org/10.1002/jcb.27257
Cai C, Huo Q, Wang X, Chen B, Yang Q (2017) SNHG16 contributes to breast cancer cell migration by competitively binding miR-98 with E2F5. Biochem Biophys Res Commun 485(2):272–278. https://doi.org/10.1016/j.bbrc.2017.02.094
Acknowledgements
We appreciate the support of all laboratory members.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
It is declared by the author that no conflict of interest exists in this paper.
Research involving human participants and/or animals
Our research protocol was approved by affiliated Tumor Hospital of Guangxi Medical University.
Informed consent
Standard written consent was obtained from each patient.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liao, S., Xing, S. & Ma, Y. LncRNA SNHG16 sponges miR-98-5p to regulate cellular processes in osteosarcoma. Cancer Chemother Pharmacol 83, 1065–1074 (2019). https://doi.org/10.1007/s00280-019-03822-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-019-03822-5