Abstract
SARS-CoV-2 has spread rapidly across the world and is negatively impacting the global human population. COVID-19 patients display a wide variety of symptoms and clinical outcomes, including those attributed to genetic ancestry. Alu retrotransposons have played an important role in human evolution, and their variants influence host response to viral infection. Intronic Alus regulate gene expression through several mechanisms, including both genetic and epigenetic pathways. With respect to SARS-CoV-2, an intronic Alu within the ACE gene is hypothesized to be associated with COVID-19 susceptibility and morbidity. Here, we review specific Alu polymorphisms that are of particular interest when considering host response to SARS-CoV-2 infection, especially polymorphic Alu insertions in genes associated with immune response and coagulation/fibrinolysis cascade. We posit that additional research focused on Alu-related pathways could yield novel biomarkers capable of predicting clinical outcomes as well as patient-specific treatment strategies for COVID-19 and related infectious diseases.
Similar content being viewed by others
Introduction
COVID-19 (coronavirus disease 2019) is a zoonotic infectious disease that primarily affects the respiratory system, caused by SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2). As of 28 December, 2020, SARS-CoV-2 has infected 79,673,754 people and is responsible for 1,761,381 deaths globally (World Health Organization; www.who.int). Pneumonia and acute respiratory distress syndrome (ARDS) are considered as the severe pulmonary manifestation of COVID-19 [1]. As SARS-CoV-2 spreads rapidly across the world, clinicians have observed a wide variety of extrapulmonary manifestations involving, at variable frequencies, multiple organs and biological systems in the human body, such as the cardiovascular, gastrointestinal, and nervous systems [1]. SARS-CoV-2 utilizes the ACE2 (angiotensin-converting enzyme 2) as a receptor for viral entry into host cells through engagement with its spike protein. The affinity of the spike protein to ACE2 is associated with the transmissibility of the virus [2] and expression levels of ACE2 are associated with susceptibility to SARS-CoV-2 [3]. The entry process requires the priming of the viral spike protein in part by TMPRSS2, a cellular surface serine protease upregulated by androgen [4]. Like SARS-CoV, SARS-CoV-2 likely downregulates ACE2 as it gains cell entry, this may directly disrupt the local equilibrium between the ACE-centered proinflammatory arms and ACE2-mediated anti-inflammatory pathways [5, 6].
General mechanisms of multi-organ damage and multi-system dysfunction observed in COVID-19 are potentially due to a combination of ubiquitous expression of ACE2 throughout the body, and the critical role ACE2 plays in maintaining the homeostasis of the renin-angiotensin-aldosterone system (RAAS) [1]. The relatively high expression of ACE2 in the cardiovascular system, especially endothelial cells of blood vessels, contributes to the thromboinflammation and hypercoagulative state prevalent in COVID-19 [7, 8]. As with many other infectious diseases, there are differential clinical manifestations and host responses among COVID-19 patients, including those having links to both biological sex and race. Preliminary analysis demonstrated that the case fatality rate is higher in males when compared to females in most countries by 1.7 times on average [9]; however, more studies are needed to confirm this observation. In the USA, while non-Hispanic black Americans comprise approximately 13.4% of the population (https://www.census.gov/quickfacts/fact/table/US/PST045219, accessed on November 30, 2020), they account for approximately 19% of deaths (https://covid.cdc.gov/covid-data-tracker/#demographics, accessed on November 18, 2020). Cultural and social determinants of health and disease are largely at play in this pandemic. Exploring the potential biological factors in addition to socioeconomic factors, such as genetic polymorphisms that underlie COVID-19 susceptibility and morbidity, is crucial in the context of disease risk, and the identification of novel personalized therapeutics.
Currently, a number of single-nucleotide polymorphisms (SNPs) in several genes and chromosome regions, such as ACE2, TMPRSS2, HLA alleles, and locus 3p21.31, have been identified to be associated with the susceptibility and outcome of COVID-19 by GWAS and exome sequencing as recently reviewed by Anastassopoulou et al. and Brest et al. [10, 11]. The initial whole-genome sequencing was completed in China and confirmed the association of HLA alleles and TMEM189-UBE2V1 region with the development of severe outcomes of COVID-19 in the Chinese population [12]. While GWAS has proven significant in identifying high-risk alleles, the design of many GWAS studies (i.e., using population-based controls instead of individuals with confirmed mild or asymptomatic COVID-19) and the intrinsic limitations of GWAS require additional efforts focused on the exploration of the biological basis for differential host response to SARS-CoV-2 [13].
Alu retrotransposons are primate-specific transposable elements that amplify in the genome through an RNA intermediate [14]. These ~300 bp long mobile genetic elements compose over 10% of the human genome [15]. Alu elements have been extremely helpful in elucidating primate evolution and demography, especially with respect to human genetic ancestry and migration patterns [16, 17]. Along the same line, numerous Alu elements are associated with SNPs that are linked to human diseases and physiological traits, including COVID-19 [18,19,20]. Indeed, an intronic Alu polymorphism in the ACE gene has already been associated with both susceptibility and morbidity of SARS-CoV-2 infection [19, 20]. A growing body of literature suggests that Alus play key regulatory roles in modulating gene expression involved in a wide range of physiological processes [21,22,23]. Knowing this, and given close ties with human demographics, it is worth considering the role Alu polymorphisms might have in the host response to SARS-CoV-2 infection, especially polymorphic Alu insertions in genes that are central to immune response and the coagulation/fibrinolysis cascade, such as ACE, PLAT, and F13B, as well as PGR which encodes for the progesterone receptor.
Here, we briefly summarize and examine known Alu polymorphisms in ACE, PGR, PLAT, F13B regarding their biological functions, roles in disease, and implications for COVID-19. The genomic information of these polymorphic Alus is provided in Table 1. Our intent is to increase awareness and spur downstream research focused on the genetic and epigenetic factors that might help explain COVID-19 susceptibility and morbidity. Such information may be critical for the development of novel biomarkers or personalized therapeutics for COVID-19 and related infectious diseases.
Methods
Selection of polymorphic Alu elements
Cardiovascular complications are common in severe cases of COVID-19 and have been ascribed to disrupted homeostasis in the renin-angiotensin-aldosterone system, the coagulation cascade, and the fibrinolysis system [1, 24]. With previous knowledge and clinical experience, we recognized that there are polymorphic Alus in the ACE, PLAT, and F13B genes that have either been confirmed or are hypothesized to influence the regulation of their corresponding products, especially in the context of cardiovascular disease and viral infection. Additionally, we include the polymorphic Alu in the PGR gene due to the known clinical effects of progesterone in regulating inflammation and cardiovascular health as well as the administration of progesterone in an ongoing COVID-19 clinical trial (ClinicalTrials.gov). We searched PubMed for all English-language original articles, reviews, and meta-analyses reporting on associations between the polymorphic Alus in the four genes and cardiovascular diseases as well as susceptibility and potential effects on outcomes of viral infection. We also searched for ongoing or completed COVID-19 clinical trials associated with ACE inhibitor, ACE receptor blocker, progesterone, and tPA at ClinicalTrials.gov.
Calculating the rate of polymorphic Alu insertions
All data of polymorphic Alu insertions were downloaded from the ALFRED database, except for Hispanic/Latino data of PGR, which was compiled from various sources including ALFRED [25,26,27] (https://www.ncbi.nlm.nih.gov/snp/, accessed on September 5). Origin of genetic ancestry (OGA) is defined within the Allele Frequency Database, ALFRED [25]. The ALFRED database was chosen due to its inclusion of multiple samplings for the same OGA. The average rate in a given OGA (Table 1) was calculated as the weighted average. All calculations and data visualizations were conducted in R.
Results
ACE
ACE (angiotensin-converting enzyme) is a ubiquitously expressed carboxypeptidase and is a central player in the hormonal regulation of the cardiovascular system, namely, the RAAS and the kallikrein-kinin system (KKS) [28]. ACE converts angiotensin I to angiotensin II, which leads to the internalization of ACE2 when in excess [29]. The antagonistic roles that ACE and ACE2 play in inflammation and anti-inflammation are crucial to physiological homeostasis and pathophysiology in the development of hypertension, renal diseases, and ARDS [6, 30,31,32,33]. The specific mechanisms implicating the RAAS and the KKS in cytokine storms during COVID-19 have been extensively reviewed [34]. Angiotensin IV, the metabolic product of angiotensin II without the participation of ACE2, modulates endothelial functions, such as plasminogen activator inhibitor-1 (PAI-1) release, which will be discussed further below [35, 36]. ACE acts as kininase to degrade bradykinin, leading to vasoconstriction, further promoting the milieu for thromboinflammation [28]. Interestingly, an intronic Alu polymorphism of ACE has been shown to directly influence the expression of the gene; individuals having a homozygous absence/deletion of the Alu (DD genotype) exhibit the highest expression of ACE, and those with homozygous insertion (II genotype) the lowest [37] (Fig. 1). The polymorphic Alu insertion is an AluYa5 element and is in complete linkage disequilibrium (LD) with an intronic variant (rs4341) and a synonymous variant (rs4343); therefore, these SNPs can be used as proxies for genotyping ACE Alu I/D (insertion/deletion) [38,39,40,41]. At the molecular level, the presence of the Alu element within intron 16 has been determined to influence the promoter activity of ACE, possibly serving as a trans-acting repressor of RNA polymerase II activity [42, 43].
Alu polymorphism within ACE intron 16 influences circulating ACE enzyme. ACE II, ID, and DD are three ACE genotypes generated by Alu insertion/deletion events. These genotypes affect the circulating level of ACE encoded by the gene. Specifically, individuals with the II genotype have the lowest circulating ACE levels whereas individuals having the DD genotype tend to have the highest-circulating ACE level. I, insertion; D, deletion
A critical observation is that the Alu polymorphism in ACE may influence the susceptibility, clinical manifestations, and outcome of COVID-19 infection multidimensionally. First, ACE can downregulate ACE2; individuals with the DD genotype would be predicted to have lower ACE2 expression, therefore, less susceptible to infection. This theory is epidemiologically supported by a negative correlation between the prevalence of COVID-19 and the ACE D-allele frequency [19]. As would be predicted, individuals of ACE Alu DD genotype have a higher propensity for developing ARDS in respiratory infections, such as Influenza A [44, 45]. Furthermore, the ACE Alu I/D polymorphism has been extensively studied across the world for a wide range of cardiovascular diseases (CVDs) and, in light of the available data, we propose closer attention paid to the ACE Alu polymorphism when considering the high prevalence of cardiovascular complications observed in COVID-19 [24]. Several studies pointed to a bleeding tendency in individuals with the II genotype, potentially through less robust coagulation, and/or enhanced fibrinolysis [46,47,48]. Studies characterizing the effects of the Alu I/D on the outcome of infectious diseases and CVDs since 2015 are provided in Supplementary Table 1. Interestingly, among patients with myocardial infarction (MI), the D-allele of ACE is associated with higher levels of IL-6, which has been associated with worse outcomes of COVID-19 [1, 49]. Considering studies of the ACE I/D polymorphism in human population genetics, the variant may in part underlie the biological factors contributing to individual race/ethnicity-related host response observed in the clinical manifestations and outcomes of COVID-19 [50].
PGR
Progesterone receptors (PGRs) are intracellular receptors that translocate to the nucleus to regulate gene expression through interacting with hormone response elements (HRE) after binding to progesterone or its derivatives [51]. Besides its well-recognized role in obstetrics and gynecology, progesterone has been studied as a potent immunomodulator in regulating many aspects of the innate and adaptive immune response [52, 53]. The relevance of progesterone with respect to vascular system function is also well-recognized. Of interest, arteries have the highest expression of PGR besides components of the female reproductive system (https://www.gtexportal.org/home/gene/ENSG00000082175). Sex-specific upregulation of endothelial mineralocorticoid receptors by endothelial progesterone receptor activation occurs in female animals with menstrual cycles [54].
Intron 7 of the PGR gene has a polymorphic AluYa5 insertion that is in complete LD with two SNPs affecting exons—V660L in exon 4 and H770H in exon 5; these polymorphisms are collectively known as PROGINS [55]. Sex steroid hormones and their downstream signaling have been hypothesized to contribute to the observed difference since they have different regulatory roles in the host immune response to COVID-19 infection [56]. Accordingly, at least one clinical trial has launched within the USA that focuses on the administration of progesterone to male patients having severe cases of COVID-19, as an attempt to mitigate mortality (ClinicalTrials.gov). In light of previous studies, we hypothesize that the Alu-related PROGINS contribute to differential host immune response. In particular, Romano and colleagues have shown that the intronic insertion of Alu decreases the responsiveness of the receptor to progestin due to a less stable transcript and proteins [57]. From the perspective of infectious diseases, PGR Alu I/D has only been investigated in the context of hepatitis E virus (HEV) and human immunodeficiency virus (HIV) [58, 59]. Regarding critical pathways related to SARS-CoV-2 infection, progesterone signaling can upregulate ACE2 in endothelial cells, thus implicating the differential response to progesterone via PGR Alu I/D in the clinical outcome of COVID-19 as it shares an intricate connection to the RAAS system [60, 61]. However, sex-related effects conferred by PGR Alu I/D remain unclear. Higher levels of progesterone in females may underly key functional differences with respect to males, and there are documented sex-specific differences in the RAAS, KKS, and fibrinolysis being affected by polymorphisms similar to those discussed herein [62]. The frequency of PROGINS does not share the trend of genetic ancestry with the Alu polymorphisms in ACE, F13B, and PLAT (Fig. 2). Nonetheless, dual characterization of Alu PROGINS and COVID-19 clinical manifestation might offer important insights into the observed ethnic and sex differences underlying the host response to SARS-CoV-2 infection.
The frequency of polymorphic Alu insertion in populations of different origin of genetic ancestry (OGA). OGAs are arranged to show an ascending frequency of Alu insertion in ACE for visualization purposes. ACE and F13B are similarly correlated with OGA, where it is the lowest in individuals with genetic ancestry associated with African populations and the highest in Asian populations. The Alu polymorphism in PLAT shares the trend with ACE and F13B but not as closely. The frequency of Alu insertion in PGR does not assume the increase observed with the polymorphisms in the other three genes. We recognize that genetic data have been misused to stigmatize ethnic groups or individuals and the usage of Alu frequencies reported herein for such purposes will not be tolerated ethically and is not scientifically justified
PLAT
PLAT encodes for tissue plasminogen activator (tPA), a serine protease found on endothelial cells that promotes fibrinolysis. It functions by converting plasminogen into plasmin, which degrades fibrin polymers, the blood clots, into D-dimers among other degradation products. The activity of tPA is inhibited by PAIs mainly secreted by endothelial cells, among other cell types and tissues. tPA is currently given as a treatment for various CVDs and ARDS in COVID-19 [63, 64], whereas elevated PAI-1 is a risk factor for both thromboembolism and atherosclerosis [65]. Both PLAT and SERPINE1 have polymorphic loci among individuals and have been studied either together or independently for predispositions and prevalence of CVDs [66]. Interestingly, levels of tPA and PAI-1 are epistatically affected by polymorphisms in the RAAS, KKS, and the fibrinolysis system [67].
Here, we focus on the structural variation caused by a polymorphic AluYa5 insertion in intron 8 of PLAT [68, 69]. The molecular effects and mechanisms have not been elucidated for this polymorphic Alu insertion; however, it was observed that individuals of the Alu II genotype have a higher “in vivo” release rate of tPA from vascular endothelial cells when compared to those with other PLAT Alu I/D genotypes [70]. Moreover, bacterial osteomyelitis patients with the PLAT Alu II genotype had lower plasma PAI-1/tPA complex levels and likely more circulating tPA [71]. This is not only very interesting in the context of bacterial infection but also potentially important for SARS-CoV-2 infection—although more active tPA mitigates the risk for developing complications of thromboembolism nature, insufficient coagulation could compromise the body’s ability to limit the pathogen locally, predisposing the individual to viremia of higher viral load and likely more severe systemic infection. In light of previous research, the PLAT Alu polymorphism, with regards to the current COVID-19 pandemic, could provide clues to the differential occurrence of coagulation and fibrinolysis dysfunction. If the effect of the Alu polymorphism was as predicted by previous studies, we expect to see individuals with the PLAT Alu DD genotype at a higher risk for developing diseases of hypercoagulative nature, such as pulmonary embolism (PE), venous thromboembolism, myocardial infarction, and stroke. In contrast, PLAT Alu II individuals would be expected to have increased risk of a variety of organ damage. It is worth noting that Ang IV (angiotensin IV) synthesis, when ACE/ACE2 ratio is higher, in the RAAS regulates the release of PAI-1 from endothelial cells; this provides the basis for potential interaction between ACE Alu I/D and PLAT Alu I/D polymorphisms [35]. Interestingly, the thrombotic syndrome in COVID-19 is similar to complement-mediated thrombotic microangiopathy (TMA) [72]. As plasminogen and plasmin can serve as a complement inhibitor and C5 convertase, respectively, the PLAT Alu I/D could also affect complement activation in COVID-19 [73]. Given the PLAT Alu I/D polymorphisms are correlated with genetic ancestry (Fig. 2); this variation may offer some biological basis for the observed patient-specific host response to SARS-CoV-2.
F13B
Plasma factor XIII is a protein in the blood coagulation cascade that stabilizes blood clots by linking fibrin monomers to fibrin polymers. FXIII is a heterotetrameric (A2B2) transglutaminase that contains two different subunits encoded by FXIIIA and FXIIIB. The A subunit is catalytically active while the B subunit protects the A subunit from degradation in circulation. Intron 10 of F13B has a polymorphic AluYa5 insertion that has been widely studied in population genetics as a genetic marker as the I-allele frequency is highly correlated with distinct population groups across the world (Fig. 2) [38, 39, 74, 75].
FXIII functions antagonistically with tPA in the blood coagulation cascade; therefore, the Alu polymorphism may prove to be essential for better prediction of the potential complications an individual might develop during COVID-19 infection for similar reasons described above. FXIIIB has been associated with the development of various bleeding disorders in recent years, including ischemic stroke (IS), venous thrombosis (VTE), coronary artery disease, and MI [76,77,78]. Of interest, the polymorphic AluYa5 insertion in F13B has been studied for conferring the risk of coronary atherosclerosis [79]. A C-to-G polymorphism in intron 11 can result in allele-specific alternative splicing that correlates with decreased FXIII albeit the significance of its level is controversial [76, 78, 80,81,82]. While it is not linked to any known SNPs, the AluYa5 insertion in intron 10 could be in LD with the C-to-G polymorphism in intron 11 given the two variants have similar racial/ethnic attributes among human populations [80]. Additional studies focused on the polymorphic Alu may elucidate additional molecular mechanisms underlying the functional regulation of FXIII. Further, the interaction between FXIIIB and proteins in the complement system, such as C3 and C1q, deserves attention for COVID-19 as reasoned in the previous section [83]. Similar to the ACE and PLAT Alu I/D polymorphism, the F13B variation observed across the human population could be an important biological factor when considering the variable individual response to COVID-19.
Discussion
SARS-CoV-2 is spreading rapidly across the globe, readily causing both pulmonary and extrapulmonary symptoms across the human population. Clinical presentations and outcomes of COVID-19 are highly variable and have been linked to race as well as sex. We reviewed studies surrounding polymorphic Alu retrotransposons in ACE, PGR, PLAT, and F13B that have been either mechanistically linked to and/or associated with clinical conditions relevant to COVID-19. In light of these studies, we posit that primate-specific Alu retrotransposons are actively influencing host physiology with respect to SARS-CoV-2 infection and are contributing to the diverse spectrum of observed COVID-19 symptoms. Moreover, variable populational rates of Alu insertion in the genes highlighted herein may provide novel insights into understanding the biological basis for the racial/ethnic discrepancies observed in the outcome of COVID-19.
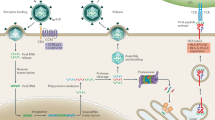
The Alu polymorphisms that we reviewed here in ACE, PGR, PLAT, and F13B constitute an example of potentially important alleles (or a combination of) that would not be identified by GWAS and non-targeted sequencing techniques. Firstly, GWAS has been mostly unsuccessful in detecting epistatic effects in humans [10, 13]. Polymorphisms in genes from the RAAS, KKS, and the fibrinolytic system, including ACE Alu I/D, are known to exert epistatic effects on the development of cardiovascular accidents [67]. When considering the available data, the Alu polymorphisms in PGR, PLAT, and F13B in addition to ACE are likely epistatically important to the homeostasis of inflammation and vascular systems and would potentially be undetected by a GWAS approach (Fig. 3). Alu polymorphisms in ACE and PGR have potential to influence the susceptibility of individuals and influence the initial viral load and tissue damage by SARS-CoV-2 by affecting the level of circulating ACE and PGR. Downregulation of ACE2 upon cell entry of SARS-CoV-2 impacts the physiological balance between ACE and ACE2, which are two essential regulatory components in the RAAS [28]. We posit that individuals with the ACE Alu DD genotype may be predisposed to complications of COVID-19 due to higher baseline ACE levels. Patients of advanced age and cardiovascular diseases or diabetes have alterations in ACE expression and Ang II signaling [84,85,86], likely rendering them more susceptible to further repercussions of ACE/ACE2 imbalance. Progesterone can upregulate ACE2 likely via transcription regulatory effects of the nuclear receptor PGR [61]. Moreover, the sensitivity to progesterone, the level of ACE, and the functions of FXIII and tPA affected by Alu structural variations can influence the spread of SARS-CoC-2 in circulation as well as predispose individuals to different tendencies of developing cardiovascular conditions (Fig. 3). SARS-CoV-2 can cause tissue and endothelial cell damage, activating the intertwined intrinsic and extrinsic blood coagulation pathways [1]. Progesterone is known to have anti-inflammatory and vasoactive effects [56, 87]. The intrinsic coagulation pathway can activate the bradykininase activity of ACE in the KKS, further promoting vasoconstriction and production of cytokines, including IL-6 [88, 89] (Fig. 3). Hemostasis is achieved through the conversion of fibrin monomers to polymers catalyzed by Factor XIII composed of FXIIIA and FXIIIB subunits in circulation. tPA and kallikrein, among other molecules, catalyze the conversion of plasminogen to plasmin, which in turn catalyzes fibrinolysis, producing D-dimer and other fibrinolysis degradation products (Fig. 3). Further, both plasmin and FXIIIB can activate the complement system in addition to their function in the fibrinolysis system [90] (Fig. 3). Dysregulated coagulation and fibrinolysis, exemplarily manifested as pulmonary embolism, cardiovascular accidents, and acute respiratory distress syndrome, are common pathophysiology observed for severe COVID-19 cases.
SARS-CoV-2 and relevant biological processes that involve ACE, PGR, FXIIIB, and tPA. SARS-CoV-2 uses ACE2 as a receptor for cell entry, facilitated by androgen-regulated TMPRSS2. SARS-CoV-2 can cause tissue and endothelial cell damages, activating the intertwined intrinsic and extrinsic blood coagulation pathways. ACE and ACE2 are two essential regulatory components in the RAAS. ACE can promote vasoconstriction through antagonizing the regulatory role of ACE2 in the RAAS and acting as a bradykininase in the KKS additionally activated by the intrinsic coagulation pathway. ACE2 is locally downregulated by SARS-CoV-2 entry and excessive ACE signaling. Progesterone can upregulate ACE2 via binding to the intracellular progesterone receptor. The result of hemostasis is the conversion of fibrin monomers to polymers catalyzed by Factor XIII, which, in circulation, is a heterotetrameric protein composed of FXIIIA and FXIIIB subunits. D-dimers constitute one of the fibrinolysis degradation products generated as plasmin catalyzes fibrinolysis. Plasminogen is the zymogen of plasmin; the transition can be facilitated by kallikrein and tPA among other molecules. Both plasmin and FXIIIB can activate the complement system in addition to their function as catalysts. The functions of proteins encoded by the ACE, PGR, PLAT, and F13B are integrally essential to the RAAS, KKS, and fibrinolytic systems; Alu variants in these genes likely contribute to the spectrum of symptoms observed in COVID-19 cases. ACE, angiotensin-converting enzyme; FXIII, Factor XIII; KKS, kallikrein-kinin system; PGR, progesterone receptor; RAAS, renin-angiotensin-aldosterone system; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane protease, serine 2; tPA, tissue plasminogen activator
Given the biological complexity of the human body, it is unlikely that a small number of SNPs in a handful of genes are independently responsible for the differential host response among the human population across the world. There are over one million copies of Alus located in the human genome and many of these mobile elements exhibit lineage-specific mobilization and epigenetic patterns that are linked to human demographics and individual natural history traits. The Alu polymorphisms presented here represent a small fraction of an individual’s global retrotransposon population, however, they likely exert epistatic efforts with other Alu and/or known non-Alu polymorphisms in the discussed systems [67]. Genetic and epigenetic mosaicisms of non-polymorphic Alus as well as polymorphic Alus located in other genes, particularly the apolipoprotein cluster (APOA1, APOC3, APOA4) and HLA loci [91,92,93], could also influence the host response and disease outcomes of COVID-19. Enhanced transcription of Alus during viral infection is a known phenomenon and the expression of Alu and other retrotransposons in human cells have been found to increase during coronavirus infection [94]. Double-stranded RNAs transcribed from Alu elements have been shown to activate antiviral innate immune signaling pathways in mitochondria through MDA5 [95]. Future efforts in characterizing the polymorphism as well as genetic and epigenetic mosaicisms of retrotransposons could reveal their physiological and pathological roles in modulating inflammation and the immune response. Such knowledge may improve our understanding of both cellular and systemic host responses to viral infections beyond COVID-19.
Conclusions
COVID-19 is a highly infectious respiratory disease caused by SARS-CoV-2, affecting multiple organs and systems in infected individuals through a complicated web of factors. In the current pandemic, clinical manifestations and outcomes of COVID-19 diverge with sex as well as differences in genetic ancestry and/or socioeconomic inequalities. Here, we present a prospective paradigm for Alu elements that has the potential to help explain differential host response to SARS-CoV-2 infection. Alu retrotransposons have been extensively researched from the perspective of human evolution and population genetics. They are well known to influence human health and disease outcomes, including those associated with genetic ancestry. Indeed, an Alu variant within the ACE gene has already been hypothesized to impact COVID-19 susceptibility and morbidity and, in light of this, several Alu variants are likely influencing host response pathways. To this end, we highlighted Alu polymorphisms in four genes encoding for ACE, PGR, tPA, and FXIIIB that are integral components of the RAAS, KKS, endocrine system, coagulation cascade, fibrinolysis, and the complement system (Fig. 3). These systems and processes are highly relevant to the development of major clinical complications of SARS-CoV-2 infection, including IL-6 levels, elevated D-dimer and fibrinogen, MI, IS, VTE, PE, and the cytokine storm as reviewed by Gupta and colleagues [1]. We posit that these Alu variants and/or the potential epistatic effects influence the clinical presentations of and host immune response to SARS-CoV-2 infection. Moreover, recently discovered epigenetic gene regulatory pathways that center on Alu elements might help explain the heterogeneity of symptoms observed across COVID-19 patients. If accurate, additional research focused on these Alu-related mechanisms could yield novel genetic markers capable of predicting the clinical outcomes as well as patient-specific treatment strategies for COVID-19.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- ACE:
-
Angiotensin-converting enzyme
- ACE2:
-
Angiotensin-converting enzyme 2
- Ang IV:
-
Angiotensin IV
- ARDS:
-
Acute respiratory distress syndrome
- COVID-19:
-
Coronavirus disease 2019
- CVDs:
-
Cardiovascular diseases
- GWAS:
-
Genome-wide association study
- HEV:
-
Hepatitis E virus
- HIV:
-
Human immunodeficiency virus
- HRE:
-
Hormone response elements
- I/D:
-
Insertion/deletion
- IL-6:
-
Interleukin-6
- IS:
-
Ischemic stroke
- KKS:
-
Kallikrein-kinin system
- LD:
-
Linkage disequilibrium
- MI:
-
Myocardial infarction
- PAI-1:
-
Plasminogen activator inhibitor-1
- PE:
-
Pulmonary embolism
- PGR:
-
Progesterone receptor
- RAAS:
-
Renin-angiotensin-aldosterone system
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- SNP:
-
Single-nucleotide polymorphism
- TMPRSS2:
-
Transmembrane Serine Protease 2
- tPA:
-
Tissue plasminogen activator
- VTE:
-
Venous thrombosis
References
Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–32.
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–20.
Yonker LM, Neilan AM, Bartsch Y, Patel AB, Regan J, Arya P, et al. Pediatric SARS-CoV-2: clinical presentation, infectivity, and immune responses. J Pediatr. 2020;227:45-52.e5.
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8.
Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B, et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11(8):875–9.
Sriram K, Insel PA. A hypothesis for pathobiology and treatment of COVID-19: the centrality of ACE1/ACE2 imbalance. Br J Pharmacol. 2020;177(21):4825–44.
Connors JM, Levy JH. Thromboinflammation and the hypercoagulability of COVID-19. J Thromb Haemost. 2020;18:1559–61.
Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004 Jun 1;203(2):631–7.
Scully EP, Haverfield J, Ursin RL, Klein SL. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat Rev Immunol. 2020;20:442–7.
Anastassopoulou C, Gkizarioti Z, Patrinos GP. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum Genomics. 2020;4:1–8.
Brest P, Refae S, Mograbi B, Hofman P, Milano G. Host polymorphisms may impact SARS-CoV-2 infectivity. Trends Genet. 2020;36(11):813–5.
Wang F, Huang S, Gao R, Zhou Y, Lai C, Li Z, et al. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discov. 2020;6(83):1–16.
Tam V, Patel N, Turcotte M, Bossé Y, Paré G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019;20:467–84.
Deininger PL, Batzer MA. Evolution of retroposons. Evol Biol. 1993;27:157–96.
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921.
Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3(5):370–9.
Cheung KH, Osier MV, Kidd JR, Pakstis AJ, Miller PL, Kidd KK. ALFRED: an allele frequency database for diverse populations and DNA polymorphisms. Nucleic Acids Res. 2000;28(1):361–3.
Payer LM, Steranka JP, Rou W, Kryatova M, Medabalimi S, Ardeljan D. Structural variants caused by Alu insertions are associated with risks for many human diseases. PNAS. 2017;114:E3984–92.
Delanghe JR, Speeckaert MM, De Buyzere ML. The host’s angiotensin-converting enzyme polymorphism may explain epidemiological findings in COVID-19 infections. Clin Chim Acta. 2020;505:192–3.
Hatami N, Ahi S, Sadeghinikoo A, Foroughian M, Javdani F, Kalani N, et al. Worldwide ACE (I/D) polymorphism may affect COVID-19 recovery rate: an ecological meta-regression. Endocrine. 2020;68(3):479–84.
Häsler J, Samuelsson T, Strub K. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell Mol Life Sci. 2007;64(14):1793–800.
Deininger P. Alu elements: know the SINEs. Genome Biol. 2011;12(12):236.
Pandey R, Mukerji M. From “JUNK” to just unexplored noncoding knowledge: the case of transcribed Alus. Brief Funct Genomics. 2011;10(5):294–311.
Nishiga M, Wang DW, Han Y, Lewis DB, Wu JC. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives. Nat Rev Cardiol. 2020;17(9):543–58.
Rajeevan H, Osier MV, Cheung K, Deng H, Druskin L, Heinzen R, et al. ALFRED: the ALelle FREquency Database. Update. 2003;31(1):270–1.
Diaz-Cueto L, Dominguez-Lopez P, Cantillo-Cabarcas J, Perez-Figueroa G, Arechavaleta-Velasco M, Arechavaleta-Velasco F. Progesterone receptor gene polymorphisms are not associated with preterm birth in a Hispanic population. Int J Gynecol Obstet. 2008;103(2):153–7.
Taylor KC, Small CM, Epstein MP, Sherman SL, Tang W, Wilson MM, et al. Associations of progesterone receptor polymorphisms with age at menarche. Horm Res Paediatr. 2010;27514:421–7.
Erdös EG, Angiotensin I. converting enzyme and the changes in our concepts through the years: Lewis K. Dahl memorial lecture. Hypertension. 1990;16(4):363–70.
Deshotels MR, Xia H, Sriramula S, Lazartigues E, Filipeanu CM. Angiotensin II mediates angiotensin converting enzyme type 2 internalization and degradation through an angiotensin II type I receptor–dependent mechanism. Hypertension. 2014;64:1368–75.
Bridget BK, A.A. NL, C. CM. Does the angiotensin-converting enzyme (ACE)/ACE2 balance contribute to the fate of angiotensin peptides in programmed hypertension? Hypertension. 2005;46(5):1097–9.
Chappel M, Ferrario C. ACE and ACE2: their role to balance the expression of angiotensin II and angiotensin-(1–7). Kidney Int. 2006;70:8–10.
Gaddam RR, Bhatia SC, Bhatia M. ACE and ACE2 in inflammation: a tale of two enzymes. Vol. 13. Inflamm Allergy Drug Targets. 2014;13(4):224–34.
Leisman DE, Deutschman CS, Legrand M. Facing COVID-19 in the ICU: vascular dysfunction, thrombosis, and dysregulated inflammation. Intensive Care Med. 2020;46(6):1105–8.
Buszko M, Park J-H, Verthelyi D, Sen R, Young HA, Rosenberg AS. The dynamic changes in cytokine responses in COVID-19: a snapshot of the current state of knowledge. Nat Immunol. 2020;21(10):1146–51.
Kerins DM, Hao Q, Vaughan DE. Angiotensin induction of PAI-1 expression in endothelial cells is mediated by the hexapeptide angiotensin IV. J Clin Invest. 1995;96(5):2515–20.
Mustafa T, Lee JH, Chai SY, Albiston AL, Mcdowall SG. Bioactive angiotensin peptides: focus on angiotensin IV. JRAAS. 2001;2(4):205–10.
Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86(4):1343–6.
Arcot SS, Shaikh TH, Kim J, Bennett L, Alegria-Hartman M, Nelson DO, et al. Sequence diversity and chromosomal distribution of “young” Alu repeats. Gene. 1995;163(2):273–8.
Batzer MA, Rubin CM, Hellmann-Blumberg U, Alegria-Hartman M, Leeflang EP, Stern JD, et al. Dispersion and insertion polymorphism in two small subfamilies of recently amplified human Alu repeats. J Mol Biol. 1995;247(3):418–27.
Glenn KL, Du Z-Q, Eisenmann JC, Rothschild MF. An alternative method for genotyping of the ACE I/D polymorphism. Mol Biol Rep. 2009 Jul;36(6):1305–10.
Abdollahi MR, Huang S, Rodriguez S, Guthrie PAI, Smith GD, Ebrahim S, et al. Homogeneous assay of rs4343, an ACE I/D proxy, and an analysis in the British Women’s Heart and Health Study (BWHHS). Dis Markers. 2008;24(1):11–7.
Mafra FFP, Gattai PP, Macedo MM, Mori MA, Araujo RC. The angiotensin-I converting enzyme insertion/deletion polymorphic element codes for an AluYa5 that down-regulates gene expression. Pharmacogenomics J. 2018;18:517–27.
Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29(4):499–509.
Onishi M, Morimoto S, Yang J, Okaishi K, Katsuya T, Shimizu M, et al. Association of angiotensin-I converting enzyme DD genotype with influenza pneumonia in the elderly. Geriatr Gerontol Int. 2002;2(1):8–15.
Marshall RP, Webb S, Bellingan GJ, Montgomery HE, Chaudhari B, McAnulty RJ, et al. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am J Respir Crit Care Med. 2002;166(5):646–50.
Kryczka KE, Płoski R, Księżycka E, Kruk M, Kostrzewa G, Kowalik I, et al. The association between the insertion/deletion polymorphism of the angiotensin-converting enzyme gene and the plasma fibrinogen level in women and men with premature coronary artery atherosclerosis. Pol Arch Intern Med. 2020;130(9):748–56.
de Carvalho SS, e Silva AC S, de Sabino A P, FCG E, Gomes KB, LMS D, et al. Influence of ACE I/D polymorphism on circulating levels of plasminogen activator inhibitor 1, D-Dimer, ultrasensitive C-reactive protein and transforming growth factor β1 in patients undergoing hemodialysis. PLoS One. 2016;11(3):e0150613.
Dušanović Pjević M, Beslac Bumbaširevic L, Vojvodic L, Grk M, Maksimović N, Damnjanović T, et al. Analysis of the association between polymorphisms within PAI-1 and ACE genes and ischemic stroke outcome after rt-PA therapy. J Pharm Pharm Sci. 2019;22(1):142–9.
Dai S, Ding M, Liang N, Li Z, Li D, Guan L, et al. Associations of ACE I/D polymorphism with the levels of ACE, kallikrein, angiotensin II and interleukin-6 in STEMI patients. Sci Rep. 2019;9(1):19719.
Watkins WS, Rogers AR, Ostler CT, Wooding S, Bamshad MJ, Brassington AE, et al. Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome Res. 2003;13:1607–18.
Beato M, Klug J. Steroid hormone receptors: an update. Hum Reprod Update. 2000;6(3):225–36.
Beagley KW, Gockel CM. Regulation of innate and adaptive immunity by the female sex hormones oestradiol and progesterone. FEMS Immunol Med Microbiol. 2003;38(1):13–22.
Kyurkchiev D, Ivanova-todorova E, Murdjeva M, Kyurkchiev S. Immunoregulation by progesterone: effects on immune cells and mesenchymal stem cells. Adv Neuroimmune Biol. 2011;1:105–23.
Faulkner JL, de JE CB. Mineralocorticoid receptor and endothelial dysfunction in hypertension. Curr Hypertens Rep. 2019;21(10):78.
Rowe S, Coughian S, McKenna N, Garrett E, Kieback D, Carney D, et al. Ovarian carcinoma-associated TaqI restriction fragment length polymorphism in intron G of the progestrone receptor G is due to an Alu sequence insertion. Cancer Res. 1995;55:2743–5.
Mauvais-Jarvis F, Klein SL, Levin ER. Estradiol, progesterone, immunomodulation, and COVID-19 outcomes. Endocrinology. 2020;161(9):1–8.
Romano A, Delvoux B, Fischer D, Groothuis P. The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone. J Mol Endocrinol. 2007;38:331–50.
López-López P, Rivero-Juarez A, Frias M, Machuca I, Caballero-Gómez J, Olivas I, et al. Mutations in the progesterone receptor (PROGINS) may reduce the symptoms of acute hepatitis E and protect against infection. Front Microbiol. 2019;10:2617.
Debes JD, Pas SD, Groothuismink ZMA, van der Ende ME, de Man RA, Boonstra A. A mutation in the progesterone receptor predisposes to HEV infection in HIV-positive patients. Liver Int. 2018;38(5):792–6.
Li Y, Zhou W, Yang L, You R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol Res. 2020;157:104833.
Chadchan SB, Maurya VK, Popli P, Kommagani R. The SARS-CoV-2 receptor, angiotensin converting enzyme 2 (ACE2) is required for human endometrial stromal cell decidualization. Biol Reprod. 2020;ioaa211:1–8. https://doi.org/10.1093/biolre/ioaa211.
Asselbergs FW, Williams SM, Hebert PR, Coffey CS, Hillege HL, Navis G, et al. The gender-specific role of polymorphisms from the fibrinolytic, renin-angiotensin, and bradykinin systems in determining plasma t-PA and PAI-1 levels. Thromb Haemost. 2006;96(4):471–7.
Hughes RE, Tadi P, Bollu PC. TPA therapy. In: StatPearls. Treasure Islang: Statpearls Publishing; 2020.
Moore HB, Barrett CD, Moore EE, Jhunjhunwala R, McIntyre RC, Moore PK, et al. Study of alteplase for respiratory failure in severe acute respiratory syndrome coronavirus 2/COVID-19: study design of the phase IIa STARS trial. Res Pract Thromb Haemost. 2020;4(6):984–96.
Vaughan D. PAI-1 and atherothrombosis. J Thromb Haemost. 2005;3(8):1879–83.
Folkert WA, Pattin K, Snieder H, Hillege HL, van Gilst WH, Moore JH. Genetic architecture of tissue-type plasminogen activator and plasminogen. Semin Thromb Hemost. 2008;34(212):562–8.
Asselbergs FW, Williams SM, Hebert PR, Coffey CS, Hillege HL, Navis G, et al. Epistatic effects of polymorphisms in genes from the renin-angiotensin, bradykinin, and fibrinolytic systems on plasma t-PA and PAI-1 levels. Genomics. 2007/01/05. 2007;89(3):362–9.
Yang-Feng TL, Opdenakker G, Volckaert G, Francke U. Human tissue-type plasminogen activator gene located near chromosomal breakpoint in myeloproliferative disorder. Am J Hum Genet. 1986 Jul;39(1):79–87.
Batzer MA, Dmingm PL. A human-specific subfamily of Alu sequences. Genomics. 1991;9(3):481–7.
Jern C, Ladenvall P, Wall U, Jern S. Gene polymorphism of t-PA is associated with forearm vascular release rate of t-PA. Arter Thromb Vasc Biol. 1999;19:454–9.
Valle-garay E, Montes AH, Corte JR, Meana A, Fierer J, Asensi V. tPA Alu (I/D) polymorphism associates with bacterial osteomyelitis; 2013. p. 208.
Merrill JT, Erkan D, Winakur J, James JA. Emerging evidence of a COVID-19 thrombotic syndrome has treatment implications. Nat Rev Rheumatol. 2020;16(10):581–9.
Barthel D, Schindler S, Zipfel PF. Plasminogen is a complement inhibitor. J Biol Chem. 2012;287(22):18831–42.
Kass DH, Aleman C, Batzer MA, Deininger PL. Identification of a human specific Alu insertion in the factor XIIIB gene. Genetica. 1994;94:1–8.
Webb GC, Coggan M, Ichinose A, Board P. g. Localization of the coagulation factor XIII B subunit gene (F13B) to chromosome bands 1q31–32.1 and restriction fragment length polymorphism at the locus. Hum Genet. 1989;81(2):157–60.
Hanscombe KB, Traylor M, Hysi PG, Bevan S, Dichgans M, Rothwell PM, et al. Genetic factors influencing coagulation factor XIII B-subunit contribute to risk of ischemic stroke. Stroke. 2015;46:2069–74.
Bereczky Z, Muszbek L. Factor XIII and venous thromboembolism. Semin Thromb Hemost. 2011;37:305–14.
Mezei ZA, Bereczky Z, Katona É, Gindele R, Balogh E. Factor XIII B subunit polymorphisms and the risk of coronary artery disease. Int J Mol Sci. 2015;16:1143–59.
Spiridonova MG, Stepanov VA, Puzyrev VP, Karpov RS. Analysis of gene complexes predisposing to coronary atherosclerosis. Genetika. 2002;38(3):383–92.
Iwata H, Kitano T, Umetsu K, Yuasa I, Yamazaki K, Kemkes-Matthes B, et al. Distinct C-terminus of the B subunit of factor XIII in a population-associated major phenotype: the first case of complete allele-specific alternative splicing products in the coagulation and fibrinolytic systems. J Thromb Haemost. 2009;7:1084–91.
Mezei ZA, Katona É, Kállai J, Bereczky Z, Molnár É, Kovács B, et al. Regulation of plasma factor XIII levels in healthy individuals; a major impact by subunit B intron K c.1952+144 C>G polymorphism. Thromb Res. 2016;148:101–6.
Balogh L, Katona É, Mezei ZA, Kállai J, Gindele R, Édes I, et al. Effect of factor XIII levels and polymorphisms on the risk of myocardial infarction in young patients. Mol Cell Biochem. 2018;448(1):199–209.
Singh S, Akhter MS, Dodt J, Volkers P, Biswas A. Identification of potential novel interacting partners for coagulation factor XIII B (FXIII-B) subunit, a protein associated with a rare bleeding disorder. Int J Mol Sci. 2019;20:2682.
Wang M, Zhang J, Jiang LQ, Spinetti G, Pintus G, Monticone R, et al. Proinflammatory profile within the grossly normal aged human aortic wall. Hypertension. 2007;50(1):219–27.
Manolis AJ, Iraklianou S, Pittaras A, Zaris M, Tsioufis K, Psaltiras G, et al. Arterial compliance changes in diabetic normotensive patients after angiotensin-converting enzyme inhibition therapy. Am J Hypertens. 2005;18(1):18–22.
Diet F, Pratt RE, Berry GJ, Momose N, Gibbons GH, Dzau VJ. Increased accumulation of tissue ACE in human atherosclerotic coronary artery disease. Circulation. 1996;11:2756–67.
Barbagallo M, Dominguez LJ, Licata G, Shan J, Bing L, Karpinski E, et al. Role of cellular calcium regulation; 2001. p. 142–7.
Du F, Liu B, Zhang S. COVID-19: the role of excessive cytokine release and potential ACE2 down-regulation in promoting hypercoagulable state associated with severe illness. J Thromb Thrombolysis. 2020:1–17.
Skurk T, Van Harmelen V, Hauner H. Angiotensin II stimulates the release of interleukin-6 and interleukin-8 from cultured human adipocytes by activation of NF-κB. Arterioscler Thromb Vasc Biol. 2004;24(7):1199–203.
Singh S, Akhter MS, Dodt J, Volkers P, Reuter A, Reinhart C, Krettler C, Oldenburg J, Biswas A. Identification of potential novel interacting partners for coagulation factor XIII B (FXIII-B) subunit, a protein associated with a rare bleeding disorder. Int J Mol Sci. 2019;20(11):2682.
Karathanasis SK. Apolipoprotein multigene family: tandem organization of human apolipoprotein AI, CIII, and AIV genes. Proc Natl Acad Sci U S A. 1985;82(19):6374–8.
Batzer MA, Stoneking M, Alegria-Hartman M, Bazan H, Kass DH, Shaikh TH, et al. African origin of human-specific polymorphic Alu insertions. Proc Natl Acad Sci USA. 1994 Dec 6;91(25):12288–92.
Kulski JK, Dunn DS. Polymorphic Alu insertions within the major histocompatibility complex class I genomic region: a brief review. Cytogenetic Genome Res. 2005;202:193–202.
Yin Y, Liu X. Exogenous coronavirus interacts with endogenous retrotransposon in human cells. Research Square. 2020:1–21. (preprint).
Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, et al. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell. 2018;172(4):797–810.e13.
Acknowledgements
We thank the University of Minnesota COVID-19 Rapid Response Clinical Grants program for funding provided to ML and PAL in support of ongoing molecular research focused on the mechanisms reviewed herein. Dr. Roxanne Larsen provided helpful comments that improved this manuscript. Figs. 1 and 3 were created with BioRender (https://biorender.com/).
Funding
The University of Minnesota COVID-19 Rapid Response Clinical Grants program for funding provided to ML and PAL in support of ongoing molecular research focused on the mechanisms reviewed herein. Discretionary funds available to PAL were used to support ML throughout the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
ML conceived the study and analyzed the data regarding the Alu polymorphisms discussed herein. ML, LS, and PAL interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Table 1.
Studies characterizing the effect that the Alu I/D within intron 16 of the ACE gene has on the outcome of various cardiovascular and infectious diseases. See Fig. 1 for depiction of the Alu polymorphism. Note that several studies highlighted within the table are underpowered and we recommend additional research having greater sample sizes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, M., Schifanella, L. & Larsen, P.A. Alu retrotransposons and COVID-19 susceptibility and morbidity. Hum Genomics 15, 2 (2021). https://doi.org/10.1186/s40246-020-00299-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40246-020-00299-9