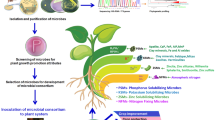
Abstract
The stalk, ear and root rot (SERR) of maize caused by Fusarium verticillioides (Fv) severely impacts crop production in tropical and subtropical regions. The aim of the present work was to screen bacterial isolates in order to find novel native biocontrol agents against Fv. A culturable bacterial collection consisting of 11,520 isolates enriched in Firmicutes and Proteobacteria was created from rhizosphere samples taken from SERR symptomatic or asymptomatic maize plants. The complete collection was screened for potential activity against Fv using a liquid antagonism assay followed by dual cultures in solid medium, selecting for 42 bacteria (Bacillus, Pseudomonas and Paenibacillus) that inhibit Fv growth (>45 %). In planta assays demonstrated that three Bacillus isolates: B. megaterium (B5), B. cereus sensu lato (B25) and Bacillus sp. (B35) displayed the highest antagonistic activity against Fv. Pot experiments performed in a greenhouse with Bacillus cereus sensu lato B25 confirmed these findings and showed a reduction of Fv disease severity and incidence on plants. Antagonistic activity analysis revealed that these strains produce glucanases, proteases or chitinases, as well as siderophores and auxins and suggests these as possible control mechanisms against Fv.
Similar content being viewed by others
Background
Maize (Zea mays L.) is one of the most important cereals grown worldwide. Maize is an important crop in Mexico due to cultural consumption habits and economic profitability. Fusarium verticillioides (Fv) (Sacc.) Nirenb. is the most commonly reported fungal species infecting maize, causing stalk, ear and root rot (SERR) of maize, and is responsible for important economic losses worldwide (Hernández-Rodríguez et al. 2008). Maize monoculture has provoked a high incidence of the disease as well as crop losses due to Fv, in Mexico’s Sinaloa state (Quintero-Benítez and Apodaca-Sánchez 2008). A consortium of four different Fusarium species (Fv, F. nygamai, F. thapsinum and F. andiyazi) belonging to the Fusarium fujikuroi species complex (FFSC) is responsible for stalk and ear rot of maize, a current problem of maize in northern Sinaloa, Mexico (Leyva-Madrigal et al. 2015). In addition to its effects on grain yield, the infection can be detrimental to grain quality (Czembor et al. 2015).
A more complete understanding of the microbial ecology and diversity associated with the maize rhizosphere could improve plant health in field crops, reduce our dependence on chemical pesticides used in agriculture, and develop efficient biological control strategies (Filion et al. 2004). The control of pathogens by sustainable agronomic practices (such as the use of biological antagonists) has recently been adopted on a commercial scale, and a number of experimental approaches are being developed (Souza et al. 2015). Plant growth-promoting rhizobacteria (PGPR) are a heterogeneous group of bacteria that can be found in the rhizosphere, at the rhizoplane or in association with roots, and can improve the extent or quality of plant growth directly or indirectly (Ahmad et al. 2008). The following genera of bacteria have been reported as PGPR: Agrobacterium, Arthrobacter, Azoarcus, Azotobacter, Azospirillum, Bacillus, Burkholderia, Caulobacter, Chromobacterium, Enterobacter, Erwinia, Flavobacterium, Klebsiella, Micrococcous, Rhizobium, Pantoea, Pseudomonas and Serratia (Bruto et al. 2014; Ahemad and Kibret 2014) which have shown potential as biocontrol agents against different fungal pathogens (Ahemad and Kibret 2014).
Seed dressing with biocontrol agents is an appropriate method to suppress plant pathogens in the spermosphere and rhizosphere (Pereira et al. 2007). In recent years, bacterial inoculants have been used to antagonize soil-borne plant pathogens such as Fv and to promote plant growth. Bacillus subtilis and Pseudomonas cepacia have been used to control root rot caused by Fv in Argentina (Cavaglieri et al. 2005b). Bacillus amyloliquefaciens or Microbacterium oleovorans can reduce the fumonisin content in harvest grains during three evaluated seasons (Pereira et al. 2011). Burkholderia spp. stimulate plant growth and suppress disease caused by Fv in maize (Hernández-Rodríguez et al. 2008), and species like Bacillus amyloliquefaciens and Enterobacter hormaechei reduce the Fv infection and fumonisin accumulation in maize kernels (Pereira et al. 2010).
Biological control may result in an effective strategy for Fv control, but it requires the development of control microorganisms that are native to the soils where maize is grown (Etcheverry et al. 2009). The introduction of a large quantity of “exotic” microorganisms may disrupt a local ecosystem and produce ecological impacts on the rhizosphere microbiota (Jackman et al. 1992). Furthermore, microbial control agents, once released, might not only repress plant pathogens, but may also affect non-target microorganisms (Pereira et al. 2009).
This work involves the massive screening of a collection of 11,520 rhizospheric bacterial isolates obtained from both symptomatic and asymptomatic SERR plants by the use of culture media that select for specific groups previously reported as Fv antagonists. Based on previous knowledge on the biology of the plant-fungus and bacteria-fungus interaction and the nature of bacteria antagonistic to Fv, we directed the screening procedure to learn if this novel strategy would allow us to enhance our chances for finding novel native rhizospheric bacterial isolates with potential biotechnological application for the control of SERR in maize. The aims of this study were to find novel potential Fv biocontrol agents, and to enhance our understanding of their plant growth-promoting and antagonistic activities.
Results
Selection of microorganisms with an antagonist effect on Fv
The bacterial collection, comprising 11,520 isolates, exhibited 95 % survival efficiency 2 months after freezing and thawing, yielding a new total of 10,944 isolates. A screen of the total bacterial collection was performed using a high throughput liquid assay (PDB) method (Figueroa-López et al. 2013). We thus selected 622 isolates showing 53–99 % Fv growth inhibition (Additional file 1: Table S1). The main bacterial genera exhibiting an Fv antagonistic effect were Bacillus (341 isolates), Enterobacter (38), Pseudomonas (23), and Lysinibacillus (13), followed by members of Acinetobacter, Agrobacterium, Anaerobranca, Aquaspirillum, Arthrobacter, Brevibacillus, Geobacillus, Klebsiella, Paenibacillus, Pantoea, Stenotrophomonas and Terribacillus (Additional file 1: Table S1).
The selection process included a second screening test based on the same principle of a conventional dual culture in solid medium in Petri dishes, but performed in 0.2 ml 96-well plates (Figueroa-López et al. 2013). Forty-two out of the 622 selected bacterial isolates from the Fusarium antagonist collection displayed 45–85 % Fv growth inhibition in PDA (Table 1 ). The criteria for selecting the most viable antagonists in this screening test were very stringent, only these 42 isolates that demonstrated on two independent experiments that three out of three replicates clearly showed an antagonistic effect against Fv were considered as viable to continue with the selection process. Most isolates belong to the genus Bacillus (34 isolates). This was represented by the most prominent groups or species: B. cereus sensu lato (21 isolates), B. megaterium (6 isolates) and B. subtilis group (5 isolates) (Table 1). Hemolytic tests were performed to discard isolates with possible pathogenic effect in humans, on the basis of their ability to produce hemolysins. Six isolates showed slight partial hemolysis or α-hemolysis (B2, Ps3, B5, B7, B12 and B13), and 8 isolates were non-hemolytic or γ-hemolytic (B4, Pa8, B9, B22, B23, B24, B25 and B35) (Table 1). The 28 isolates exhibiting β-hemolysis (total hemolysis) were discarded. The remaining selected isolates were used for the sterile in planta assay, to test for their antagonistic behavior in the presence of the host plant.
In planta assays
Sterile sand assay
Thirteen isolates were applied to maize plants and tested as PGPRs. Isolate B13 showed a significant increase (60 %) in root volume in Cebu hybrid as compared to the control inoculated with Fv (Fig. 1a). Isolates Ps3, B5, B25 and B35 significantly increased root volume in the Garañón hybrid as compared to the control inoculated with Fv (Fig. 1b).
In planta antagonistic assays 45 days after seed emergence in two white maize hybrids inoculated with 14 partial or non-hemolytic bacterial isolates and Fv. a Root volume (Cebú hybrid), comparing a water control against the addition of the bacterial isolate. b Percentage of disease severity (Cebú hybrid), comparing a water control against the addition of the bacterial isolate. c Root volume (Garañón hybrid), comparing a water control against the addition of the bacterial isolate. d Percentage of disease severity (Garañón hybrid), comparing a water control against the addition of the bacterial isolate. CTL refers to the fungus control (plant plus Fv). Letters preceding a number indicate the genus of that particular isolate: B refers to Bacillus, Ps is Pseudomonas and Pa is Paenibacillus. Identical letters appearing above bars indicate no significant differences, while different letters indicate significant differences (Tukey P ≥ 0.05)
In the Cebú white maize hybrid, isolates B5 (47 %), B13 (62 %) and B25 (53 %) significantly reduced Fv disease severity compared to control plants inoculated with Fv (100 %) (Fig. 1c). Conversely, isolates B5, B25 and B35 reduced the Fv disease severity by 45–50 % in the Garañón hybrid, as compared to the Fv-treated control (Fig. 1d). An additional experiment was run in parallel to evaluate the possible effect of bacteria on root volume. This confirmed that those isolates causing a difference in the presence of Fv did not induce significant differences in root volume when applied alone to the seed, as compared to untreated plants (data not shown).
Although Ps3, B5, B13, B25 and B35 showed plant growth promoting activity on in vitro plant assays, the B5 and B25 strains decreased disease severity on both hybrids. We selected B25 to conduct further greenhouse tests because it was γ-hemolytic and this reduces the possibility of this strain to become pathogenic to humans.
Greenhouse experiments
Fusarium natural inoculum in soil was ~2 × 106 c.f.u./g and it increased after soil inoculation in treatments with F. verticillioides P03 to ~6 × 107 c.f.u./g. Incidence of Fusarium root and stalk rot was significantly reduced by strain B25 (Table 2). Stalk rot measured as percentage of severity decreased significantly when inoculated with B25 compared to both controls: untreated and Fv inoculated, while root rot severity decreased significantly with respect to the untreated control treatment but not to the Fv treated control plants (Table 2).
Plant growth-promoting and antagonistic traits of bacterial isolates tested in planta
To explore which mechanisms of growth promotion could be involved in Fv control, we examined different antagonistic traits in the isolates evaluated in the maize antagonistic assays (Table 3). Phosphate solubilization was detected in isolates B4, B5, Pa8, B12, B13 and B23. IAA production was shown only for Pa8, which produced 40 µmol/l of auxin-like compounds. Siderophores were produced by isolates Ps3, B4, B5, B7, B12, B13, B22, B24 and B25. Protease activity was present in B4, B5, B7, B12, B22, B24 and B25. Chitinase activity was displayed by isolates B13, B23, B24 and B25. All isolates exhibited glucanase activity except for B12 and B23 (Table 3).
Discussion
The effect of a pathogen on native microbial communities has previously been studied in diseases affecting crops besides maize, such as citrus (Trivedi et al. 2010; Araujo et al. 2002), conifers (Filion et al. 2004), wheat (McSpadden Gardener and Weller 2001), potato (Reiter et al. 2002), and avocado (Yang et al. 2001). These studies demonstrate that phytopathogens affect both endophytic and rhizospheric bacteria populations, and that some bacterial populations may assist the plant in preventing disease symptoms, by inhibiting phytopathogen growth.
The aim of our work was to find bacterial isolates that control Fv growth. Our approach, a fast high-throughput screening liquid antagonistic assay coupled to a conventional dual-culture antagonism assay, allowed identifying such isolates that could be potential Fv antagonists. The rationale for using both screening methods was that the first screening could select not only for diffusible substances produced by the bacterial isolates affecting Fv growth but also for organisms that could affect fungal growth only when they enter in contact with the fungus, the dual-culture antagonism assay should then confirm which of those selected isolates have the best antagonistic effect against Fv before entering in contact with the fungus. The main reason for executing the screening in this order was that the liquid screening is less time-consuming than the dual-culture antagonism assay. It is possible that by performing the screening in this order we had missed a few isolates that are effective controlling Fv growth by entering in contact with the fungus. Using different types of antagonism assays during a large screening like the one performed in this work should allow selecting for isolates that employ different biocontrol strategies.
Massive screening assays designed to find potential antagonistic bacteria against fungal phytopathogens have limitations proper of the specific test. The initial screening step performed on PDB was biased to favor Fv growth. Although preliminary tests (Figueroa-López et al. 2013) allowed us to learn that most bacterial isolates would be able to grow well in PDB, we found bacteria that either did not show growth at all (>2.5 %), or did exhibit visually poor growth in PDB (>1 %) (data not shown). These bacteria probably will not be able to produce enough of the substances that may inhibit Fv growth and will not reach a threshold that will allow them to exhibit an antagonistic response. Thus, it is possible that our initial screening assay may have left aside a few potential isolates that were unable to perform well under these assay conditions.
Different species of Bacillus represented more than half of the identified Fv antagonistic isolates (Additional file 1: Table S1), as well as the most prominent bacterial populations in this study. Different species belonging to all other genera that showed fungal inhibition in liquid assay (Additional file 1: Table S1) including Acinetobacter (Magnin-Robert et al. 2007), Arthrobacter (Cavaglieri et al. 2005a), Enterobacter (van Dijk and Nelson 1997; Gopalakrishnan et al. 2011), Klebsiella (Lynch 1990), Lysinibacillus (Trivedi et al. 2011), Paenibacillus (Nielsen and Sorensen 1997), Pantoea (Babalola 2010) and Pseudomonas (Gorlach-Lira and Stefaniak 2009) are reported to possess antagonistic traits against diverse pathogens.
Pseudomonas putida and P. fluorescens inhibited Fv growth in the solid medium assay (Table 1). Different Pseudomonas spp. produce a broad array of lytic enzymes, antibiotics, cyanide, siderophores and antifungal compounds (Nagarajkumar et al. 2004; Weller et al. 2007; Gorlach-Lira and Stefaniak 2009) which can all inhibit pathogens including Fv.
The Bacillus genus is able to produce many secondary metabolites with antifungal effects on diverse plant pathogens (Raaijmakers and Mazzola 2012). Cavaglieri et al. (2005a) demonstrated the antagonistic effect of ten Bacillus isolates (including B. cereus) against Fv with growth inhibition percentages ranging from 28 to 78 %; by comparison, our study revealed percentages between 45 and 85 % in solid medium (Table 1). Bacillus species use diverse mechanisms that may inhibit this fungal pathogen (Ongena and Jacques 2008) including nutrient competition (Kamilova et al. 2005), production of antifungal lipopeptides (Nihorimbere et al. 2012), or production of lytic enzymes such as chitinases that can degrade the fungal cell wall as a means to avoid fungal hyphal extension (Kishore et al. 2005). The stringent conditions used in the dual-plate assay reduced significantly the amount of isolates from 622 to only 42. Nevertheless, we cannot discard the possibility that by doing this we had missed some isolates that could be effective in the next selection steps. Time and space constraints for performing in planta assays, either in growth chambers or greenhouses, constitute important factors for selecting fewer isolates to work with in plant species such as maize.
In vitro testing of B. subtilis in maize roots and kernels has previously been reported to inhibit both Fv growth and the production of fumonisin B1 (Cavaglieri et al. 2004). Nevertheless, results obtained in vitro are not necessarily reproducible when the host plant is included in its tripartite interaction with the fungus and the bacterium. This is probably due to the plant exudates, which could affect the fungus and/or bacteria in different ways (Fan et al. 2012). Furthermore, different results may be obtained when the potential bacterial antagonist is placed under field conditions and other biotic or abiotic factors are considered (Egamberdiyeva 2007). On the other hand, satisfactory results using endophytic B. subtilis in maize causes a reduction in mycotoxin production and a decrease in Fv colonization (Bacon et al. 2001). Bacillus cereus increased grain yield by 43.8 % in maize (Tilak and Reddy 2006).
Application of bacteria to seeds (bacterization) has been widely used for the biological control of soil-borne plant pathogens that affect many host plants (Cavaglieri et al. 2005a). In this work, we used plant-based experiments to evaluate 13 native isolates of the maize rhizosphere that are capable of inhibiting Fv growth in vitro. Our results suggest a beneficial response in growth promotion (measured as root volume), as well as on Fv disease severity between some isolates and the white maize hybrids tested (Fig. 1). Bacillus megaterium (B5) and B. cereus sensu lato (B25) reduced Fv disease severity in both white maize hybrids tested. Bacillus megaterium has been reported to act as a biocontrol agent of Fusarium crown and root rot of tomato (Omar et al. 2006).
We selected B25 to conduct greenhouse pot experiments based on its lack of hemolytic activity and ability to decrease Fv disease severity in both white maize hybrids tested (Garañón and Cebú) under sterile in vitro conditions. Many human pathogenic bacteria produce soluble proteins that can lyse erythrocytes (Herlax and Bakas 2002), the B25 strain was γ-hemolytic (no-hemolysis) suggesting that it is not harmful to human health. B25 was assayed under non-sterile conditions and a third white maize hybrid (DK2038 from Dekalb) commonly employed in northern Sinaloa, Mexico.
B25 reduced the disease incidence and in most cases disease severity confirming the in vitro results and suggesting a good capability to establish well in the rhizosphere and compete against other rhizospheric microorganisms present in the soil. This effect could be due to the different plant growth promotion activities that B25 possesses (Table 3).
Another focus of our study was plant growth-promoting or antagonistic traits used by isolates to inhibit Fv growth and decrease disease severity in maize plants. We investigated these traits by performing several tests and found that isolates use different mechanisms (Table 3). Phosphate solubilizing bacteria, including Bacillus species (Kumar et al. 2012), can play an important role in plant nutrition, by increasing phosphorus uptake in plants (Rodríguez et al. 2007). The siderophores produced by PGPRs are capable of inhibiting root pathogens by creating limiting iron conditions in the rhizosphere. The Bacillus isolates B5, B13 and B25 reported in this work are siderophore producers and potential biocontrol agents (Table 3), which are able to reduce the disease severity in maize plants (Table 2; Fig. 1). One Bacillus subtilis strain has been reported to produce bacilibactin and itoic acid, as well as siderophores, and is able to reduce Fusarium wilt incidence in pepper (Yu et al. 2011). On the other hand, isolates B5 and B25 were observed to exhibit protease activity. These antifungal proteins are responsible for inhibition against diverse pathogens including Fv in this work, and other fungi including F. oxysporum, F. solani, P. ultimum and Rhizoctonia solani (Chang et al. 2008; Gao et al. 2008). In this study, isolates of B. cereus sensu lato (B24 and B25), B. subtilis group (B13) and Bacillus megaterium (B23) showed chitinase activity, suggesting that it could be a potential trait involved in Fv growth control for these isolates (Bressan and Fontes-Figueiredo 2010). Production of chitinases by Bacillus was demonstrated to cause a reduction in F. graminearum infection in wheat (Shali et al. 2010). Chitin and glucan are the main structural components of the fungal cell wall (Yang et al. 2004), and most isolates (including B5, B13, B25 and B35) produced glucanases. The observed chitinase and glucanase activities suggest that they may be implicated in the inhibition and reduction of disease severity by all isolates producing these enzymes.
Some bacteria may share similar repertoires of hydrolytic enzymes or antagonistic traits when tested in vitro, but the results may differ when the other participants in this interaction (i.e. the plant and the fungus) intervene. This is suggested by the observation that the pairs B4 and B5 or B24 and B25 share similar activities, despite the fact that no Fv control was exerted by B4 or B24. Although the underlying reasons are currently not understood, our research group is currently conducting work in parallel to elucidate the mechanisms used by these bacteria to exert biocontrol in planta.
Special consideration was given to the discovery of native bacterial isolates in this work, since such biocontrol agents could be well suited to edaphic and climatological conditions from a specific region. Additionally, co-existence for many years with the natural soil microbiota should provide native microorganisms with competitive advantages compared to exotic species. The research presented here was aimed at identifying native biocontrol agents and bacteria capable of exerting biocontrol against Fv, as well as providing valuable information on the agriculturally natural conditions of these bacterial populations in maize plants. Only cultivable microorganisms can be able to develop bio-fertilizers against crops diseases (Jha et al. 2010) and the native microorganism are promising alternative. Our findings with Fv antagonistic assays in vitro and in planta have been recently confirmed in field trials (Lizárraga-Sánchez et al. 2015) and the efficacy of the isolates have been proved in order to exploit them as potential biocontrol agents suitable for widespread use in the large extensions of maize sown in Sinaloa, Mexico.
Conclusions
In this study we analyzed a large number of microorganisms using an easy and time-saving in vitro screening method that allowed us to find a potential biocontrol agent in a short amount of time. We suggest for future studies concerning the selection of bacterial antagonists against fungal plant pathogens that before designing a protocol for massive screening assays, decisions should be taken based on: (1) the best available knowledge on the biology of the specific plant-fungal interaction both in vitro and under field conditions; (2) the possible nature and source of the isolates; (3) the best isolation and assay culture media; (4) the relative abundance of the potential antagonists in the rhizosphere microbiota; and (5) as many factors as possible to plan the screening protocol. Screening of a collection of 11,520 rhizospheric bacteria allowed identifying three Bacillus isolates (B5, B25, B35) as potential antagonists to inhibit maize rots caused by Fv. These strains produce lytic enzymes such as glucanases, proteases or chitinases, as well as siderophores and auxins and suggests these as possible control mechanisms against Fv. Bacillus cereus sensu lato B25 was selected to conduct greenhouse pot assays that showed a reduction of Fv disease severity and incidence on plants confirming the potential of this isolate to control Fv in maize. B25 strain was effective in controlling Fv possibly due to the several PGPR traits that the bacterium possesses. B25 is currently being studied to develop a novel biological product based on spore production (Martínez-Álvarez et al. 2016) to reduce Fusarium stalk, ear and root rots in maize fields.
Methods
Sample collection
A total of fifty maize rhizosphere samples were collected from five locations in Sinaloa state (Mexico): (1) Serrano; (2) Alhuey; (3) 18 de Diciembre; (4) Casa Blanca; and (5) La Trinidad. These fields showed symptoms of plant damage by the fungus Fv. Plants were sampled in five paired groups, each pair consisting of one symptomatic plant and one asymptomatic plant grown side by side. The five samples from each condition and location were homogenized together and stored at room temperature. 3–4 kg of bulk soil were removed from the stem base of each plant. Each of the five sampling points differed by planting day and by maize hybrids. Microbiological analyses were conducted to confirm SERR symptomatology, and Fv was isolated from SERR symptomatic plants in selective media.
Sample processing
Soil particles adhering to the roots (rhizospheric soil) were collected and microorganisms were then isolated by serial dilutions. Four different culture media were prepared in 100 mm-diameter Petri dishes to enrich for specific taxonomic groups: Luria Bertani (LB) medium was used to enrich for Bacillus isolates (Cavaglieri et al. 2005a); Actinomycetes Isolation Agar (AIA) was used for Actinomycetes isolates (Bressan and Fontes-Figueiredo 2007); King B Agar (KBA) was used for Pseudomonas (Cavaglieri et al. 2004); and Man, Rogosa and Sharpe (MRS) medium was used for lactic acid bacteria (De Man et al. 1960). Colonies were collected from LB, KBA and MRS media after 24 h growth and from AIA medium after 48–72 h at 25 °C. A bulk soil sub-sample (500 g) was used for nutrient and physicochemical soil analyses.
Microorganism collection and viability test
To generate the maize rhizospheric culturable bacterial collection, 288 isolates were “picked” and arranged in three 96-well plates from each specific culture medium. This was performed for each composite sample with the five symptomatic-rhizosperic samples, as well as the five asymptomatic rhizosperic samples per each maize field. This yielded 1152 isolates from each of the ten composite rhizospheric sample points; the complete collection therefore contained 11,520 isolates. Isolates were cryopreserved in triplicate at −70 °C, using LB containing 15 % glycerol (v/v) according to Pasarell and McGinnis (1992). Frozen stocks were made and grown at 25 °C in rotary shaker at 200 rev/min in 2 ml 96-well plates containing 1.5 ml liquid medium, for either 24 h (LB, KBA and MRS media) or 72 h (AIA medium). The isolate was considered non-viable if no visible growth was observed after thawing. Plates containing bacterial pellets from viable isolates were stored at −70 °C until processing for DNA extraction.
Molecular identification of bacteria using 16S rDNA
Bacterial DNA was extracted with the DNeasy® Blood & Tissue Kit (Qiagen; CA, USA). The primers F2C (5′-AGAGTTTGATCATGGCTC-3′) and C (5′-ACGGGCGGTGTGTAC-3′) (Shi et al. 1997) were used to amplify 16S rDNA. PCR reactions were carried out in 96-well plates. The 25 μl PCR mixture contained 10 ng of DNA template, 1X reaction buffer, 10 pmol of each primer, 10 µmol/l of each deoxynucleoside triphosphate (dNTP), and 1 U of Taq DNA polymerase (Invitrogen; Carlsbad, CA, USA). The PCR conditions included an initial denaturation step at 95 °C (4 min); 32 cycles of denaturation at 95 °C (1 min) followed by annealing at 60 °C (5 min) and extension at 72 °C (1.5 min); and a final step at 72 °C (5 min). The PCR was performed using a MyCycler thermal cycler (BioRad; CA, USA). Products were visualized by 1 % agarose (w/v) gel electrophoresis in 0.5 X Tris–acetate-EDTA (TAE) buffer and stained with ethidium bromide. PCR products were purified with a QIAquick PCR Purification kit (Qiagen; CA, USA) and quantitated using a Nanodrop 2000 UV–Vis spectrophotometer (Thermo Scientific). The U1 primer (5′-CCAGCAGCCGCGGTAATACG-3′) (Lu et al. 2000) internal to the F2C/C amplified PCR product was used for sequencing with an ABI 3730 XL automated sequencer at the National Laboratory of Genomics (LANGEBIO; Irapuato, Mexico). Isolates were identified by sequence comparison against the Ribosomal Database Project (RDP) and GenBank databases using the BLASTN search algorithm (http://blast.ncbi.nlm.nih.gov). The sequences were then compared on the basis of identity percentage, E-value and Match score, using the default parameters from the RDP seq match tool.
Screening of Fv antagonists
The antagonism selection assay was performed to analyze all viable bacterial specimens from the collection, according to (Figueroa-López et al. 2013). Selection was performed in two steps. First, a liquid antagonism assay using potato dextrose broth (PDB) was performed. Briefly, this consisted of growing the isolate and the fungus together in 2 ml 96-well plates and quantifying the fungal biomass by staining the chitin residues of the fungal cell wall with the wheat germ agglutinin lectin coupled to a fluorophore (WGA Alexa Fluor 488 conjugate), which was then measured using a multimodal fluorescence detector (Beckman, DTX800). The selection criterion was arbitrarily set at >50 % fungal growth inhibition. The bacterial isolates derived from this screen were assayed in a second selection step using regular dual antagonism assays with 0.2 ml of potato dextrose agar (PDA) medium in 96 well plates to confirm the effect observed in the first assay. In this second assay, the selection criterion was set for isolates to display ≥45 % fungal growth inhibition in all three replicates.
Blood hemolytic assay
Hemolysis tests were performed in order to discard isolates that could be pathogenic to humans. Bacterial isolates were grown in 15 ml tubes containing 5 ml of LB at 25 °C for 24 h in a rotary shaker at 250 rev/min. One ml of bacterial culture was taken and transferred to a 1.5 ml tube, centrifuged twice at 16,800g for 5 min, and the resulting supernatant was transferred to a new tube. 5 mm-diameter wells were made in blood agar plates with a cork borer, and 50 µl of supernatant were aliquoted into the wells. The plates were then stored at 37 °C for 24 h. Complete β-hemolysis was observed as a clear zone around the well in the blood agar medium, indicating complete breakage of erythrocytes. Conversely, partial α-hemolysis was observed as a dark-green coloration around the well, indicating the partial damage of erythrocytes. Bacteria with γ-hemolysis do not exhibit any alteration of color or opacity in the medium, indicating an absence of hemolysis (Forbes et al. 2002).
In planta antagonistic assays
Sterile sand assay
Two types of white maize hybrid seeds, Cebú and Garañón (Asgrow), were used for the in planta antagonism assay. Seeds were surface-sterilized prior to bioassays by placing them in 0.75 % sodium hypochlorite (w/v) at 52 °C for 20 min, followed by three copious washes with sterile distilled water for 5 min each (Daniels 1983). This methodology yielded 98–100 % seed germination with 1–3 % seed contamination. For this reason, seeds were pre-germinated on Komada’s Fusarium-selective medium (Komada 1975), and seeds with no symptoms of fungal growth (i.e. Fv-free) were selected, whereas those presenting contamination were eliminated. Bacterial isolates were grown in 15 ml tubes containing 5 ml of LB medium at 25 °C for 24 h at 250 rev/min, and an optical density (OD) of 1.0 at 595 nm was used to calculate the colony-forming units (c.f.u./ml) after plating. Maize seeds were soaked in bacterial suspensions containing 1.5 × 108 c.f.u./ml for 20 min. Three seeds were planted per sterile polypropylene container (similar to a Magenta box) containing 200 g of wet sterile sand. Bacterial-treated seeds were transferred to sand inoculated with Fv isolate P03 2 days before sowing, and control untreated seeds were placed in sterile sand containing Fv (which was added at a concentration of 1 × 105 conidia/g of sand). Nine plants per control or treatment were evaluated in three experimental sets containing three seeds each. The experiment was evaluated 45 days after seed emergence. Root volume was measured according to Burdett (1979) and disease severity was evaluated as described in Cumagun et al. (2009). An additional experiment was performed as a control to confirm that bacteria did not cause any detrimental effects to root volume; this experiment used a set of bacterial-treated seeds compared to an untreated seed control (receiving water) grown in sterile sand under the same conditions as listed above (data not shown).
Greenhouse assay
Fv isolate P03 was reactivated on PDA plates by incubation at 25 °C for 14 days. Three mycelial plugs (7 mm diameter) were transferred to sterile plastic bags containing 100 g of sterile cracked maize, hydrated with 40 ml of sterile distilled water and incubated at 25 °C for 14 days (Leyva-Madrigal et al. 2015). Sixty-five grams of the maize/fungus mix was then added to 1 kg styrofoam pots containing a mixture of non-sterile vermiculite:non-sterile soil (1:1 v/v). Fungal inoculum concentration (c.f.u./g) was estimated using the “massive stamping drop plate” method (Corral-Lugo et al. 2012) in Nash–Snyder agar plates. To inoculate plants with bacteria, root systems of five-days-germinated plantlets (hybrid Dk2038 from Dekalb®) were submerged for 10 min in a bacterial suspension (5 × 108 c.f.u/ml) of B25, blot dried and then planted in a pot. One plant was placed per pot (one replicate). A completely randomized design with 5 replicates per treatment was used. The following treatments were included: untreated control (untreated maize seeds); strain B25 (maize seeds treated with B. cereus B25 without Fv P03); control Fv (untreated maize seeds containing Fv P03); strain B25 + Fv (maize seeds treated with B. cereus B25 and Fv P03).
Pots were kept in greenhouse conditions at 28 ± 2 °C with a natural photoperiod for 40 days, where they were watered two times a week with distilled water. Disease incidence was evaluated by visual assessment and reported as positive if the presence of any signs of disease was observed. Visual disease symptoms on roots were assessed according to the severity scale reported by Soonthornpoct et al. (2000). Data were converted to a disease index score using the formula reported by Asran and Buchenauer (2003).
Characterization of functional plant growth-promoting and/or antagonistic traits of bacterial isolates
Plate screening assays were used to investigate plant growth-promoting and antagonistic traits. Auxin production was evaluated using Salkowsky’s reagent (Loper and Schroth 1986). Briefly, single colonies were grown in LB broth for 24 h and the supernatants were treated with Salkowsky’s reagent, according to Bric et al. (1991). After several minutes, IAA production was identified by a color change in the supernatant. Isolates were streaked and screened for phosphate-solubilizing ability on Pikosvkaya’s agar (Pikosvkaya 1948). After incubation for 1 week at 25 °C, the presence of a clear zone around the bacterial colony was considered positive for phosphate solubilizing. The chitinase assay was performed on colloidal chitin agar medium according to (Shanmugaiah et al. 2008), and chitinase activity was identified by the formation of a clear zone around the bacterial cells after 5 days of growth. In vitro β-1, 4-endoglucanase activity was assayed using carboxy-methyl cellulose (CMC; Cat 419273, Sigma Chemicals Company; St. Louis, MO, USA) as the substrate. Single colonies were grown in LB broth for 48 h at 30 °C. Two hundred µl of cell-free supernatant was placed in 5 mm-diameter wells (previously made using a cork borer) in 1 % CMC agar plates, and incubated for 24 h at 30 °C. The formation of a clear zone around a well, resulting from β-1, 4-endoglucanase activity, was revealed by adding 5 ml of Congo red 1 % w/v for 15 min. Subsequently, the Congo red dye was removed and 5 ml NaCl 2 mol/l was added for 15 min to eliminate the excess dye, and to visualize the formation of clear zones (Teather and Wood 1982). Siderophore production was determined after 1 week of incubation in chrome azurol S (CAS) agar. The CAS blue solution for this assay was prepared according to Schwyn and Neilands (1987). Pure isolates were pricked onto CAS agar plates using sterile toothpicks and incubated at 25 °C for 2 weeks in the dark, and the assay was performed in triplicate. Colonies with yellow/orange zones were considered to be siderophore-producing strains. Protease activity was tested in skimmed milk agar (SMA) with commercially available non-fat milk, according to Jones et al. (2007). The strains were streaked onto SMA, and plates were incubated for 24 h at 30 °C. Protease activity was identified by the formation of a clear zone around the bacterial colonies. All assays were performed in triplicate for each bacterial isolate tested.
Statistical analyses
A completely randomized experimental design was used for plant experiments. The obtained severity scale values were evaluated by a normality test, using the Shapiro Will test and a Bartlett’s test to confirm variance homogeneity. Data were parametric, and severity scale data were subjected to statistical analysis of variance (ANOVA) to detect differences between treatments. All percentage values were previously converted to arcsine [√(x %/100) + 0.5] for data normalization and to proceed with the analysis of variance (Dughetti and García 2004). Mean comparisons were made using Tukey’s and Duncan’s tests; all statistical tests were conducted at a probability level of P ≤ 0.05. All analyses were performed using the Statistical Analysis System 9.0 software (SAS Institute; Cary, NC).
Abbreviations
- Fv :
-
Fusarium verticillioides
- SERR:
-
stalk, ear and root rot
- LB:
-
Luria Bertani
References
Ahemad M, Kibret M (2014) Mechanisms and applications of plant growth promoting rhizobacteria: current perspective. J King Saud Univ Sci 26(1):1–20. doi:10.1016/j.jksus.2013.05.001
Ahmad F, Ahmad I, Khan MS (2008) Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res 163(2):173–181. doi:10.1016/j.micres.2006.04.001
Araujo WL, Marcon J, Maccheroni W, van Elsas JD, van Vuurde JWL, Azevedo JL (2002) Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol 68(10):4906–4914. doi:10.1128/aem.68.10.4906-4914.2002
Asran MR, Buchenauer H (2003) Pathogenicity of Fusarium graminearum isolates on maize (Zea mays L.) cultivars and relation with deoxynivalenol and ergosterol contents. J Plant Dis Prot 110:209–219
Babalola OO (2010) Beneficial bacteria of agricultural importance. Biotechnol Lett 32(11):1559–1570. doi:10.1007/s10529-010-0347-0
Bacon CW, Yates IE, Hinton DM, Meredith F (2001) Biological control of Fusarium moniliforme in maize. Environ Health Perspec 109:325–332
Bressan W, Fontes-Figueiredo JE (2007) Efficacy and dose–response relationship in biocontrol of Fusarium disease in maize by Streptomyces spp. Eur J Plant Pathol 120(3):311–316. doi:10.1007/s10658-007-9220-y
Bressan W, Fontes-Figueiredo JE (2010) Chitinolytic Bacillus spp. isolates antagonistic to Fusarium moniliforme in maize. J Plant Pathol 92(2):343–347. doi:10.4454/jpp.v92i2.175
Bric JM, Bostock RM, Silverstonef SE (1991) Rapid in situ assay for indole acetic acid production by bacteria immobilized on titrocellulose membrane. Appl Environ Microbiol 57:535–538
Bruto M, Prigent-Combaret C, Muller D, Moënne-Loccoz Y (2014) Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci Rep 4:6261. doi:10.1038/srep06261
Burdett AN (1979) A nondestructive method for measuring the volume of intact plant parts. Can J For Res 9(1):120–122. doi:10.1139/x79-021
Cavaglieri L, Passone A, Etcheverry M (2004) Screening procedures for selecting rhizobacteria with biocontrol effects upon Fusarium verticillioides growth and fumonisin B1 production. Res Microbiol 155(9):747–754. doi:10.1016/j.resmic.2004.06.001
Cavaglieri L, Andrés L, Ibáñez M, Etcheverry M (2005a) Rhizobacteria and their potential to control Fusarium verticillioides: effect of maize bacterisation and inoculum density. Antonie Van Leeuwenhoek 87(3):179–187. doi:10.1007/s10482-004-3193-z
Cavaglieri L, Orlando J, Rodríguez MI, Chulze S, Etcheverry M (2005b) Biocontrol of Bacillus subtilis against Fusarium verticillioides in vitro and at the maize root level. Res Microbiol 156(5–6):748–754. doi:10.1016/j.resmic.2005.03.001
Chang W-T, Hsieh C-H, Hsieh H-S, Chen C (2008) Conversion of crude chitosan to an anti-fungal protease by Bacillus cereus. World J Microbiol Biotechnol 25(3):375–382. doi:10.1007/s11274-008-9901-5
Corral-Lugo A, Morales-García YE, Pazos-Rojas LA, Ramírez-Valverde A, Martínez-Contreras RD, Muñoz-Rojas J (2012) Cuantificación de bacterias cultivables mediante el método de “Goteo en Placa por Sellado (o estampado) Masivo”. Rev Colomb Biotecnol 14:147–156
Cumagun CJ, Ramos JS, Dimaano AO, Munaut F, Hove F (2009) Genetic characteristics of Fusarium verticillioides from corn in the Philippines. J Gen Plant Pathol 75(6):405–412. doi:10.1007/s10327-009-0199-4
Czembor E, Stępień Ł, Waśkiewicz A (2015) Effect of environmental factors on Fusarium species and associated mycotoxins in maize grain grown in poland. PLoS ONE 10(7):e0133644. doi:10.1371/journal.pone.0133644
Daniels BA (1983) Elimination of Fusarium moniliforme from corn seed. Plant Dis 67:609–611. doi:10.1094/PD-67-609
De Man JC, Rogosa M, Sharpe ME (1960) A medium for the cultivation of Lactobacilli. J Appl Bacteriol 23(1):130–135. doi:10.1111/j.1365-2672.1960.tb00188.x
Dughetti AC, García C (2004) Prueba de la tolerancia de distintos materialesde cebolla de días largos, al ataque de Delia spp. (Diptera: Anthomyidae). In: Paper presented at the XXVII Conferencia Congreso Argentino de Horticultura, VI Reunión Científica de la Cebolla del Mercosur. I Jornadas de Productos Frutihortícolas para una Alimentación Saludable, San Luis, 21–24 semtiembre
Egamberdiyeva D (2007) The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Appl Soil Ecol 36(2–3):184–189. doi:10.1016/j.apsoil.2007.02.005
Etcheverry M, Scandolara A, Nesci A, Vilas Boas Ribeiro MS, Pereira P, Battilani P (2009) Biological interactions to select biocontrol agents against toxigenic strains of Aspergillus flavus and Fusarium verticillioides from maize. Mycopathologia 167(5):287–295. doi:10.1007/s11046-008-9177-1
Fan B, Carvalhais LC, Becker A, Fedoseyenko D, von Wiren N, Borriss R (2012) Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol 12(116):1471–2180. doi:10.1186/1471-2180-12-116
Figueroa-López AM, Cordero-Ramírez JD, Quiroz-Figueroa FR, Maldonado-Mendoza IE (2013) A high-throughput screening assay to identify bacterial antagonists against Fusarium verticillioides. J Basic Microbiol 54(S1):125–133. doi:10.1002/jobm.201200594
Filion M, Hamelin RC, Bernier L, St-Arnaud M (2004) Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl Environ Microbiol 70(6):3541–3551. doi:10.1128/aem.70.6.3541-3551.2004
Forbes BA, Sahm DF, Weissfeld AS (2002) Bailey and Sco***tt’s diagnostic microbiology, 11th edn. Mosby, St. Louis MO
Gao X-A, Ju W-T, Jung W-J, Park R-D (2008) Purification and characterization of chitosanase from Bacillus cereus D-11. Carbohydr Polym 72(3):513–520. doi:10.1016/j.carbpol.2007.09.025
Gopalakrishnan S, Humayun P, Kiran B, Kannan I, Vidya M, Deepthi K, Rupela O (2011) Evaluation of bacteria isolated from rice rhizosphere for biological control of charcoal rot of sorghum caused by Macrophomina phaseolina (Tassi) Goid. World J Microbiol Biotechnol 27(6):1313–1321. doi:10.1007/s11274-010-0579-0
Gorlach-Lira K, Stefaniak O (2009) Antagonistic activity of bacteria isolated from crops cultivated in a rotation system and a monoculture against Pythium debaryanum and Fusarium oxysporum. Folia Microbiol 54(5):447–450. doi:10.1007/s12223-009-0062-1
Herlax V, Bakas LS (2002) Aplicaiones terapéuticas de toxinas líticas formadoras de poros: potencialidades de alfa-hemolisina de Escherichia coli. Medicina 62:66–72
Hernández-Rodríguez A, Heydrich-Pérez M, Acebo-Guerrero Y, Velazquez-del Valle MG, Hernández-Lauzardo AN (2008) Antagonistic activity of Cuban native rhizobacteria against Fusarium verticillioides (Sacc.) Nirenb. in maize (Zea mays L.). Appl Soil Ecol 39(2):180–186. doi:10.1016/j.apsoil.2007.12.008
Jackman SC, Lee H, Trevors JT (1992) Survival, detection and containment of bacteria. Microb Releases 1:125–154
Jha CK, Patel D, Rajendran N, Saraf M (2010) Combinatorial assessment on dominance and informative diversity of PGPR from rhizosphere of Jatropha curcas L. J Basic Microbiol 50(3):211–217. doi:10.1002/jobm.200900272
Jones BV, Sun F, Marchesi JR (2007) Using skimmed milk agar to functionally screen a gut metagenomic library for proteases may lead to false positives. Lett Appl Microbiol 45(4):418–420. doi:10.1111/j.1472-765X.2007.02202.x
Kamilova F, Validov S, Azarova T, Mulders I, Lugtenberg B (2005) Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ Microbiol 7(11):1809–1817. doi:10.1111/j.1462-2920.2005.00889.x
Kishore GK, Pande S, Podile AR (2005) Biological control of late leaf spot of peanut (Arachis hypogaea) with chitinolytic bacteria. Phytopathology 95(10):1157–1165. doi:10.1094/PHYTO-95-1157
Komada H (1975) Development of a selective medium for quantitative isolation of Fusarium oxysporum from natural soil. Rev Plant Protec Res 8:114–125
Kumar P, Dubey RC, Maheshwari DK (2012) Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol Res 167(8):493–499. doi:10.1016/j.micres.2012.05.002
Leyva-Madrigal KY, Larralde-Corona CP, Apodaca-Sánchez MA, Quiroz-Figueroa FR, Mexia-Bolaños PA, Portillo-Valenzuela S, Maldonado-Mendoza IE (2015) Fusarium species from the Fusarium fujikuroi species complex involved in mixed infections of maize in Northern Sinaloa, Mexico. J Phytopathol 163(6):486–497. doi:10.1111/jph.12346
Lizárraga-Sánchez GJ, Leyva-Madrigal KY, Sánchez-Peña P, Quiroz-Figueroa FR, Maldonado-Mendoza IE (2015) Bacillus cereus sensu lato strain B25 controls maize stalk and ear rot in Sinaloa, Mexico. Field Crops Res 176:11–21. doi:10.1016/j.fcr.2015.02.015
Loper JE, Schroth MN (1986) Influence of bacterial sources of indole-3-acetic acid on root elongation of sugar beet. Phytopathology 76(4):386–389. doi:10.1094/Phyto-76-386
Lu JJ, Perng CL, Lee SY, Wan CC (2000) Use of PCR with universal primers and restriction endonuclease digestions for detection and identification of common bacterial pathogens in cerebrospinal fluid. J Clin Microbiol 38(6):2076–2080
Lynch JM (1990) Beneficial interactions between micro-organisms and roots. Biotechnol Adv 8(2):335–346. doi:10.1016/0734-9750(90)91069-S
Magnin-Robert M, Trotel-Aziz P, Quantinet D, Biagianti S, Aziz A (2007) Biological control of Botrytis cinerea by selected grapevine-associated bacteria and stimulation of chitinase and β-1,3 glucanase activities under field conditions. Eur J Plant Pathol 118(1):43–57. doi:10.1007/s10658-007-9111-2
Martínez-Álvarez JC, Castro-Martínez C, Sánchez-Peña P, Gutiérrez-Dorado R, Maldonado-Mendoza IE (2016) Development of a powder formulation based on Bacillus cereus sensu lato strain B25 spores for biological control of Fusarium verticillioides in maize plants. World J Microbiol Biotechnol. doi:10.1007/s11274-015-2000-5
McSpadden Gardener BB, Weller DM (2001) Changes in populations of rhizosphere bacteria associated with take-all disease of wheat. Appl Environ Microbiol 67(10):4414–4425. doi:10.1128/aem.67.10.4414-4425.2001
Nagarajkumar M, Bhaskaran R, Velazhahan R (2004) Involvement of secondary metabolites and extracellular lytic enzymes produced by Pseudomonas fluorescens in inhibition of Rhizoctonia solani, the rice sheath blight pathogen. Microbiol Res 159(1):73–81. doi:10.1016/j.micres.2004.01.005
Nielsen P, Sorensen J (1997) Multi-target and medium-independent fungal antagonism by hydrolytic enzymes in Paenibacillus polymyxa and Bacillus pumilus strains from barley rhizosphere. FEMS Microbiol Ecol 22(3):183–192. doi:10.1016/s0168-6496(96)00089-x
Nihorimbere V, Cawoy H, Seyer A, Brunelle A, Thonart P, Ongena M (2012) Impact of rhizosphere factors on cyclic lipopeptide signature from the plant beneficial strain Bacillus amyloliquefaciens S499. FEMS Microbiol Ecol 79(1):176–191. doi:10.1111/j.1574-6941.2011.01208.x
Omar I, O’Neill TM, Rossall S (2006) Biological control of Fusarium crown and root rot of tomato with antagonistic bacteria and integrated control when combined with the fungicide carbendazim. Plant Pathol 55(1):92–99. doi:10.1111/j.1365-3059.2005.01315.x
Ongena M, Jacques P (2008) Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol 16(3):115–125. doi:10.1016/j.tim.2007.12.009
Pasarell L, McGinnis MR (1992) Viability of fungal cultures maintained at −70 °C. J Clin Microbiol 30(4):1000–1004
Pereira P, Nesci A, Etcheverry M (2007) Effects of biocontrol agents on Fusarium verticillioides count and fumonisin content in the maize agroecosystem: impact on rhizospheric bacterial and fungal groups. Biol Control 42(3):281–287. doi:10.1016/j.biocontrol.2007.05.015
Pereira P, Nesci A, Etcheverry M (2009) Impact of two bacterial biocontrol agents on bacterial and fungal culturable groups associated with the roots of field-grown maize. Lett Appl Microbiol 48(4):493–499. doi:10.1111/j.1472-765X.2009.02558.x
Pereira P, Nesci A, Castillo C, Etcheverry M (2010) Impact of bacterial biological control agents on fumonisin B1 content and Fusarium verticillioides infection of field-grown maize. Biol Control 53(3):258–266. doi:10.1016/j.biocontrol.2010.02.001
Pereira P, Nesci A, Castillo C, Etcheverry M (2011) Field studies on the relationship between Fusarium verticillioides and maize (Zea mays L.): effect of biocontrol agents on fungal infection and toxin content of grains at harvest. Int. J Agron 2011:7. doi:10.1155/2011/486914
Pikosvkaya RI (1948) Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiologia 17:362–370
Quintero-Benítez JA, Apodaca-Sánchez MA (2008) Las pudriciones de tallos en el maíz y su manejo en Sinaloa. In: MC C (ed) Jornada de manejo sustentable del cultivo del maíz. Memoria de Capacitación. Fundación Produce Sinaloa, Gobierno del Estado de Sinaloa, Sinaloa
Raaijmakers JM, Mazzola M (2012) Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol 50(1):403–424. doi:10.1146/annurev-phyto-081211-172908
Reiter B, Pfeifer U, Schwab H, Sessitsch A (2002) Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol 68(5):2261–2268. doi:10.1128/aem.68.5.2261-2268.2002
Rodríguez H, Fraga R, Gonzalez T, Bashan Y (2007) Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. In: Velázquez E, Rodríguez-Barrueco C (eds) First international meeting on microbial phosphate solubilization, vol 102. Developments in plant and soil sciences. Springer Netherlands, pp 15–21. doi:10.1007/978-1-4020-5765-6
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160(1):47–56. doi:10.1016/0003-2697(87)90612-9
Shali A, Ghasemi S, Ahmadian G, Ranjbar G, Dehestani A, Khalesi N, Motallebi E, Vahed M (2010) Bacillus pumilus SG2 chitinases induced and regulated by chitin, show inhibitory activity against Fusarium graminearum and Bipolaris sorokiniana. Phytoparasitica 38(2):141–147. doi:10.1007/s12600-009-0078-8
Shanmugaiah V, Mathivanan N, Balasubramanian N, Manoharan PT (2008) Optimization of cultural conditions for production of chitinase by Bacillus laterosporous MML2270 isolated from rice rhizosphere soil. Afr J Biotechnol 7(16):2562–2568
Shi T, Reeves RH, Gilichinsky DA, Friedmann EI (1997) Characterization of viable bacteria from Siberian permafrost by 16S rDNA sequencing. Microb Ecol 33:169–179. doi:10.1007/s002489900019
Soonthornpoct P, Trevathan LE, Ingram D (2000) The colonization of maize seedling roots and rhizosphere by Fusarium spp. in Mississippi in two soil types under conventional tillage and notillage systems. Phytoprotection 81:97–106
Souza Rd, Ambrosini A, Passaglia LMP (2015) Plant growth-promoting bacteria as inoculants in agricultural soils. Gen Mol Biol 38:401–419
Teather RM, Wood PJ (1982) Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol 43(4):777–780
Tilak BR, Reddy BS (2006) Bacillus cereus and B. circulans—novel inoculants for crops. Curr Sci 90(5):642–644
Trivedi P, Duan Y, Wang N (2010) Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl Environ Microbiol 76(11):3427–3436. doi:10.1128/aem.02901-09
Trivedi P, Spann T, Wang N (2011) Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microbial Ecol 62(2):324–336. doi:10.1007/s00248-011-9822-y
van Dijk K, Nelson EB (1997) Inactivation of seed exudate stimulants of Pythium ultimum sporangium germination by biocontrol strains of Enterobacter cloacae and other seed-associated bacteria. Soil Biol Biochem 30(2):183–192
Weller DM, Landa BB, Mavrodi OV, Schroeder KL, De La Fuente L, Bankhead SB, Allende Molar R, Bonsal RF, Mavrodi DV, Thomashow LS (2007) Role of 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. in the defense of plant roots. Plant Biol 9(1):4–20. doi:10.1055/s-2006-924473
Yang CH, Crowley DE, Menge JA (2001) 16S rDNA fingerprinting of rhizosphere bacterial communities associated with healthy and Phytophthora infected avocado roots. FEMS Microbiol Ecol 35:129–136. doi:10.1111/j.1574-6941.2001.tb00796.x
Yang J, Kharbanda PD, Mirza M (2004) Evaluation of Paenibacillus polymyxa pkb1 for biocontrol of Pythium disease of cucumber in a hydroponic system. Acta Hortic 635:59–66
Yu X, Ai C, Xin L, Zhou G (2011) The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur J Soil Biol 47(2):138–145. doi:10.1016/j.ejsobi.2010.11.001
Authors’ contributions
AMFL conducted most of the trials, research and drafted the manuscript. JDCR participated in conducting the in vitro plant assays. JCMA and LSGJ conducted the greenhouse assays. MLM, RFG and CCM participated in the design and analysis of some experiments, CCM helped with the plate assays. IEMM conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank F. R. Quiroz-Figueroa for technical help with sequence analysis. AMFL and JDCR received Ph.D. fellowships from the Consejo Nacional de Ciencia y Tecnología (CONACyT) (Mexico) and SIP-IPN. We thank Dr. Brandon Loveall of Improvence for English proofreading of the manuscript. Financial support for this project was provided by the Fundación Produce Sinaloa (2009–2013) and the Secretaría de Investigación y Posgrado del Instituto Politécnico Nacional (SIP-IPN) (2009–2014).
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional file
40064_2016_1780_MOESM1_ESM.docx
Additional file 1: Table S1. Isolates showing >50 % Fv growth inhibition obtained from the large-scale liquid antagonism assay, and the corresponding name of the 42 isolates (Name column) selected for their antagonistic activity in solid medium (see Table 1).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Figueroa-López, A.M., Cordero-Ramírez, J.D., Martínez-Álvarez, J.C. et al. Rhizospheric bacteria of maize with potential for biocontrol of Fusarium verticillioides . SpringerPlus 5, 330 (2016). https://doi.org/10.1186/s40064-016-1780-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40064-016-1780-x