Abstract
Objective
Early identifying sepsis patients who had higher risk of poor prognosis was extremely important. The aim of this study was to develop an artificial neural networks (ANN) model for early predicting clinical outcomes in sepsis.
Methods
This study was a retrospective design. Sepsis patients from the Medical Information Mart for Intensive Care-III (MIMIC-III) database were enrolled. A predictive model for predicting 30-day morality in sepsis was performed based on the ANN approach.
Results
A total of 2874 patients with sepsis were included and 30-day mortality was 29.8%. The study population was categorized into the training set (n = 1698) and validation set (n = 1176) based on the ratio of 6:4. 11 variables which showed significant differences between survivor group and nonsurvivor group in training set were selected for constructing the ANN model. In training set, the predictive performance based on the area under the receiver-operating characteristic curve (AUC) were 0.873 for ANN model, 0.720 for logistic regression, 0.629 for APACHEII score and 0.619 for SOFA score. In validation set, the AUCs of ANN, logistic regression, APAHCEII score, and SOFA score were 0.811, 0.752, 0.607, and 0.628, respectively.
Conclusion
An ANN model for predicting 30-day mortality in sepsis was performed. Our predictive model can be beneficial for early detection of patients with higher risk of poor prognosis.
Similar content being viewed by others
Introduction
Sepsis, as a syndrome of organ dysfunction induced by a dysregulated response to infection, was one of major causes leading to high mortality and poor clinical outcomes in intensive care unit(ICU) [1, 2]. Studies reported that the short-term and long-term mortality of sepsis varied from 20 to 50% [3,4,5]. Hence, early identifying sepsis patients who had higher risk of poor prognosis was extremely important for physicians so they can do some intervention and timely managements to improve the clinical outcomes [6].
Artificial neural networks (ANN), as a type of machine learning algorithm, have been applied widely for medical researches [7,8,9]. One study with a total of 21,892 cases showed that ANN model had a good performance for predicting 14-day hospital readmission with pneumonia [10]. Another recent research on cancer demonstrated that ANN model was capable of simultaneously predicting the multiple co-occurring symptoms including the risk of pain, psychological disorders and lack of well-being [11]. In the COVID-19 pandemic, scientific researchers in Brazil applied the ANN model to easily make daily and cumulative forecasts for cases and deaths so that government officials and medical agencies could do actions more agilely and reliably [12].
In the present study, we aimed to explore the capability of ANN model in predicting clinical outcomes in sepsis based on the publicly accessible database of Medical Information Mart for Intensive Cart III (MIMIC-III).
Methods
Database and patients
MIMIC-III database is a US-based critical care public database. Clinical and laboratory data associated with 53,423 age ≥ 16 patients from 2001 to 2012 and 7870 neonates from 2001 to 2008 admitted in ICU were documented [13]. The database mainly included charted events such as demographics, vital signs, laboratory tests, vital status, medications, image reports, and clinical outcomes.
All patients with sepsis (ICD9 code: 99,591) in MIMIC-III (version 1.4) were enrolled in this study. Exclusion criteria included as follows: patients with missing > 5% individual data and age less than 18.
Data extraction
From the MIMIC-III database, the following general variables were extracted for the first 24 h after ICU admission: age at the time of hospital admission, gender, admission type, marital status, ethnicity, ICU department, comorbidities (renal disease, coronary artery disease (CAD), diabetes, and hypertension), sequential organ failure assessment (SOFA) score and acute physiology and chronic health evaluation (APACHEII) score. The length of stay (LOS) in ICU and in-hospital mortality were also collected.
Clinical and laboratory variables which were recorded within 24 h after admission were also extracted including systolic blood pressure (SBP), diastolic blood pressure (DBP), heart rate (HR), respiratory rate (RR), white blood cells (WBC), neutrophils, lymphocytes, sodium, chloride, platelet (PLT), red cell volume distribution width (RDW), mean corpusular volume (MCV), hematocrit, glucose, prothrombin time (PT), partial thrombin time (PTT), albumin, alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin, urea nitrogen, creatinine, lactate, total calcium, and anion gap. NLR is defined as the ratio of neutrophils to lymphocytes. Multiple multivariable imputations were utilized for addressing missing data to maximize statistical power and minimize bias.
Statistical analysis
Descriptive statistics included as follow: proportions and frequencies were used for categorical variables, while medians, mean (SD), and interquartile ranges (IQRs) were used for continuous variables. Chi-squared test or Mann–Whitney U test were utilized for the comparison between the survivor group and the nonsurvivor group.
First, univariable analysis was applied for identifying variables which were significantly different between the two groups. Then, those variables were enrolled to construct the predictive model by multivariable logistic regression. At last, the receiver-operator characteristic (ROC) analysis for predicting 30-day mortality was performed and the area under the curve (AUC) estimates were calculated. The analyses of accuracy, sensitivity, and specificity were also done for evaluating the predictive performance of different models. The best threshold values of variables were confirmed by the Youden Index (sensitivity+specificity-1). The value of each variable with the maximum Youden Index was the best threshold value.
Statistical analysis was performed by using SPSS software (version 26). A p value of < 0.05 was considered as statistically significant.
ANN model
For our ANN model, a multilayer perception with back propagation algorithm was the applied architecture [14, 15]. The basic structure of ANN had three layers including the input layer, the hidden layer and the output layer (Fig. 2). The variables which showed significant differences between the survivor group and nonsurvivor group by using univariate analysis were enrolled in the input layer. In Fig. 2, our ANN was composed with 1 input layer consisting of 12 nodes, 1 hidden layer consisting of 6 nodes, and 1 output layer consisting of 2 nodes.
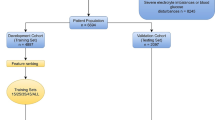
The study population was categorized into the training set (n = 1689) and the validation set (n = 1176) was based on the ratio of 6:4 by simple randomization using R software function of set.seed (), respectively. We applied an oversampling algorithm method to deal with the imbalance between training set and validation set [16]. The training set was utilized to construct models and the validation set was used to test the predictive performance of the models (Table 2). The predictive performance of ANN was analyzed by averaging the 30-day mortality from the fivefold cross-validation [11]. In addition, the average accuracy, sensitivity, and specificity were calculated. The predictive performances of ANN, logistic regression, APACHEII, and SOFA scores were compared for training set and validation set were compared. ANN model was performed with PyTorch (version1.2.0).
Results
General characteristics of sepsis in MIMIC-III
At first, a total of 5403 patients with sepsis were enrolled. Based on the exclusion criteria, 2874 patients were included in our study (Fig. 1). The 30-day mortality was 29.8%. The median age of the cohort was 67, and males accounted for 55.7% in total. Among marital status, the proportions of divorced, married, single and widow individuals were 6.8%, 44.5%, 28.4%, and 15.4%, respectively. Most of the patients were white (72.7%). 96% of patients were admitted in emergency and more than a half were transferred in MICU (65.9%). Among comorbidities, the proportions of renal disease, CAD, diabetes and hypertension were 8.4%, 15.9%, 5.4%, and 37.9%, respectively. The median scores of SOFA and APACHE in the cohort were 2 and 14, respectively. The median days of LOS in ICU and hospital were 3 and 8, respectively (Table 1).
Baseline characteristics of training and validation tests
Table 2 demonstrated the general characteristics of training and validation. Except for diabetes (P = 0.021), there was no significant difference in other variables including age (P = 0.213), gender (P = 0.994), DBP (P = 0.310), SBP (P = 0.763), HR (P = 0.122), RR (P = 0.148), renal disease (P = 0.930), CAD(P = 0.542), hypertension (P = 0.774), PLT (P = 0.849), AST (P = 0.303), sodium(P = 0.931), glucose (P = 0.194), chloride (P = 0.510), MCV (P = 0.096), ALT(P = 0.420), neutrophils (P = 0.144), urea nitrogen(P = 0.617), PTT(P = 0.886), hematocrit (P = 0.355), PT (P = 0.949), anion gap (P = 0.070), RDW (P = 0.612), lymphocytes (P = 0.063), WBC (P = 0.089), NLR (P = 0.088), total calcium (P = 0.381), lactate (P = 0.790), albumin (P = 0.169), creatinine (P = 0.893), total bilirubin (P = 0.743), APACHEII (P = 0.581), SOFA (P = 0.671), LOS in hospital (P = 0.386) and 30-day mortality (P = 0.153).
Multivariable logistic regression analysis
In Table 3, significant differences were showed in variables including age (P < 0.001), AST (P < 0.001), MCV (P = 0.001), ALT (P < 0.001), urea nitrogen (P < 0.001), PTT (P < 0.001), PT (P < 0.001), RDW (P < 0.001), lactate (P < 0.001), albumin (P < 0.001) and total bilirubin (P < 0.001) between two groups in the training set.
11 variables were enrolled in multivariable logistic regression analysis and 9 variables were identified as independent factors associated with 30-day mortality (Table 4): age(odds ratio (OR) 1.030,95% CI 1.020–1.039), AST(OR 1.000, 95% CI 1.000–1.001), urea nitrogen(OR 1.008,95% CI 1.004–1.013), RDW(OR 1.161, 95% CI 1.098–1.227), lactate(OR = 1.189, 95% CI 1.115–1.268), albumin(OR 0.581, 95% CI 0.447–0.708), total bilirubin(OR 1.059, 95% CI 1.029–1.091), PT(OR 1.031, 95% CI 1. 010–1.052) and PLT(OR 0.999, 95% CI 0.998–1.000).
ANN model development
The main structures of artificial neural networks were illuminated in Fig. 2. 11 variables including age, AST, MCV, ALT, urea nitrogen, PTT, PT, RDW, lactate, albumin and total bilirubin which showed significant differences between two groups were selected for the input layer. The output layer was 30-day hospital mortality. In Fig. 3, normalized importance of all 11 variables were demonstrated. The top four significant variables were albumin (100.00%), PT (85.73%), RDW (82.81%), and lactate (76.75%).
Predictive performance of different models in Training set and Validation set In Table 5, predictive performance of ANN, logistic regression, APAHCEII and SOFA scores for training set and validation set were demonstrated. In training set, the accuracies of the four models were 0.866, 0.711, 0.615, and 0.574, respectively (P < 0.001). The sensitivities were 0.850, 0.662, 0.569, and 0.619, respectively (P < 0.001). The specificities were 0.410, 0.337, 0.367 and 0.413, respectively (P = 0.029). The area under the ROC curve (AUC) of ANN, LR, APACHEII and SOFA scores were 0.873, 0.720, 0.629 and 0.619, respectively (P < 0.001). In validation set, the accuracies of the four models were 0.735, 0.722, 0.401, and 0.609, respectively (P = 0.272). The sensitivities were 0.624, 0.604, 0.333, and 0.416, respectively (P = 0.197). The specificities were 0.772, 0.744, 0.841, and 0.788, respectively (P = 0.095). The AUCs of ANN, LR, APACHEII, and SOFA scores were 0.811, 0.752, 0.607, and 0.628, respectively (P = 0.002).
Comparison of the predictive performances in different models Figure 4 showed the ROCs of ANN, LR, APACHEII, and SOFA scores for training set (A) and validation set (B), which showed that the ANN model had the highest ROCs in both training set and validation set. In Table 6, AUCs of ANN, LR, APACHEII and SOFA scores between training set and validation set were compared. ANN model showed the significant difference (P < 0.001), while no significant difference was found in logistic regression (P = 0.067), APACHEII score (P = 0.174) and SOFA score (P = 0.350).
The receiver operating characteristic curves of ANN, LR, SOFA, APACHEII in predicting 30-day mortality in sepsis. 4A: Training set; 4B: Validation set. ANN artificial neural networks, SOFA sequential organ failure assessment, APACHE acute physiology and chronic health evaluation, LR logistic regression
Discussion
In our study, an ANN model for predicting 30-day mortality in sepsis was performed. To our best knowledge, it was the first study for investigating the performance of ANN model in predicting short-term outcomes in sepsis based on MIMIC-III database.
Compared to LR model, ANN was good at dealing with nonlinear correlation in different analyses and also had a superiority in analysis of variables with sophisticated correlations [17]. One Korean study clarified that a total of 1260 bacteremia episodes were identified in 13,402 patients and ANN model had a better performance in early detection of bacteremia, with an AUC of 0.729 and a sensitivity of 0.810 [18]. Another study concluded that when ANN model was applied to the prediction of individual episodes of apnea and hypopnea in people with obstructive sleep apnea syndrome, it had both good specificity and sensitivity [19]. Our study showed that ANN model with an AUC of 0.811 was significantly superior to compared to LR, SOFA score and APACHEII score.
Four most important variables including albumin, PT, RDW, and lactate were identified in our ANN model. Accumulating evidence demonstrated those four variables were associated with clinical outcomes in sepsis [20,21,22].
Albumin, as the main protein which can balance capillary membrane permeability and plasma osmotic pressure, was identified to be associated with occurrence and clinical outcomes in sepsis [23]. One study clarified that low serum albumin levels (< 29.2 g/L) was an independent risk factor for 28-day mortality in sepsis [24]. Furthermore, the daily changes of albumin were significantly linked with mortality during the ICU stay in sepsis patients [25]. Another retrospective study concluded that in sepsis, the probability of survival decreased by 63.4% when serum albumin was ≤ 2.45 g/dl on admission, and by 76.4% when the lowest serum albumin during hospitalization was ≤ 1.45 g/dl [26].
Previous research illuminated that coagulation function on ICU admission was associated with mortality in sepsis [21]. In septic shock, survival curve analysis demonstrated a higher of PT/INR (> 0.16) had significantly higher risk in 28-day mortality compared with a lower level (< 0.16) [27]. One recent COVID-19 study found that non-survivors with sepsis had higher level of PT and APTT [28]. In sepsis, due to infection and activated innate immune system, coagulation will be activated, leading to sepsis associated coagulopathy with over-consumption of coagulation factors [29].
RDW, as a parameter for evaluating in the size of circulating red blood cells, was to be identified as a predictive indicator in different disorders [30,31,32,33]. A sepsis study with a total of 566 patients with overall mortality of 29% demonstrated that higher RDW was independently associated with 28-day mortality [34]. Another study investigated the association between RDW and in-hospital mortality in sepsis and found that RDW had good predictive performance with the AUC of 0.867 [35]. In a study on sepsis-induced acute respiratory distress syndrome, cox regression model showed that RDW was also an independent prognostic marker [36].
Lactate was reported as a predictor for the risk of death in all patients with or without sepsis [37]. Hyperlactatemia was more frequent in septic shock and was associated with a lower survival rate [38]. A prospectively research with a cohort of 1233 adults in UK showed that a lactate ≥ 2 mmol/L was associated with an increase in mortality and identified patients with suspicion of sepsis who had the highest risk of in-hospital mortality [39]. Lactate showed the similar prognostic accuracy for mortality in adults with sepsis compared to that of SOFA [4]. The current research proved that in polymicrobial sepsis, lactate could promote macrophage high mobility group box-1(HMGB1) lactylation/acetylation and release exosome, leading to disrupted endothelium integrity and increased vascular permeability [40].
In our study, we performed a predictive model for 30-day mortality in sepsis using ANN. Our predictive model can be beneficial for the early detection of patients with higher risk of poor prognosis. When those patients with higher risk of mortality are identified, physicians can do some intervention and timely managements in order to improve the clinical outcomes. Although the predictive model couldn’t help guide ICU management, it may be more relevant to target short-term outcomes including respiratory failure or vasopressor initiation within 48 h which could impact disposition decisions.
Some limitations should be stated in our study. First, the MIMIC-III public database included data before 2012, while the new definition of Sepsis-3.0 was published in 2016. Differences in the definition of sepsis in different phrases should be considered when applying our ANN model. Second, due to a high percentage of missing values in MIMIC-III, not all the variables which may affect the clinical outcomes in sepsis were included and analyzed. Some variables including the percentage of patients that received antibiotics, and the timing of such were not analyzed, which may confound the outcome of 30-day mortality. Third, the ANN model was applied to perform this study. Whether other prediction models of machine learning have better predictive performance than the ANN model should be further investigated. Fourth, our study constructed a predictive ANN model for 30-day mortality in sepsis. The primary outcome was 30-day mortality and patients with out-of-hospital mortality within 30 days might be missed. Fifth, we only investigated the 30-day mortality as the main outcome in the study. Other outcomes including complications and long-term prognosis were not investigated. In the future, further studies including more samples and longer follow-up should be conducted to help explore how to improve the clinical outcomes in sepsis.
Conclusion
In our study, an ANN model for predicting 30-day mortality in sepsis was performed. The predictive model can be beneficial for the early detection of patients with higher risk of poor prognosis.
Availability of data and materials
The datasets used and/or analyzed during the present study were availed by the corresponding author on reasonable request.
Abbreviations
- SOFA:
-
Sequential organ failure assessment
- APACHE:
-
Acute physiology and chronic health evaluation
- LOS:
-
Length of stay
- ICU:
-
Intensive care unit
- CAD:
-
Coronary artery disease
- SBP:
-
Systolic blood pressure
- DBP:
-
Diastolic blood pressure
- HR:
-
Heart rate
- RR:
-
respiratory rate
- WBC:
-
White blood cells
- PLT:
-
Platelet
- RDW:
-
Red cell volume distribution width
- PT:
-
Prothrombin time
- PTT:
-
Partial thrombin time
- ALT:
-
Alanine aminotransferase
- INR:
-
International normalized ratio
- AST:
-
Aspartate aminotransferase
- MCV:
-
Mean corpuscular volume
- LOS:
-
Length of stay
- ICU:
-
Intensive care unit
- IQR:
-
Interquartile ranges
- CI:
-
Confidential interval
- OR:
-
Odds ratio
- AUC:
-
Area under the curve
- ROC:
-
Receiver-operator characteristic
- HMGB1:
-
High mobility group box-1
Referencess
Zhang Z, Hong Y. Development of a novel score for the prediction of hospital mortality in patients with severe sepsis: the use of electronic healthcare records with LASSO regression. Oncotarget. 2017;8(30):49637–45.
Li S, Hu X, Xu J, Huang F, Guo Z, Tong L, Lui KY, Cao L, Zhu Y, Yao J, et al. Increased body mass index linked to greater short- and long-term survival in sepsis patients: a retrospective analysis of a large clinical database. Int J Infect Dis. 2019;87:109–16.
Yang Y, Liang S, Geng J, Wang Q, Wang P, Cao Y, Li R, Gao G, Li L. Development of a nomogram to predict 30-day mortality of patients with sepsis-associated encephalopathy: a retrospective cohort study. J Intensive Care. 2020;8:45.
Liu Z, Meng Z, Li Y, Zhao J, Wu S, Gou S, Wu H. Prognostic accuracy of the serum lactate level, the SOFA score and the qSOFA score for mortality among adults with Sepsis. Scand J Trauma, Resusc Emerg Med. 2019;27(1):51.
Zheng R, Pan H, Wang J, Yu X, Chen Z, Pan J. The association of coagulation indicators with in-hospital mortality and 1-year mortality of patients with sepsis at ICU admissions: a retrospective cohort study. Clinica chim Acta Int J Clin Chem. 2020;504:109–18.
Shen Y, Huang X, Zhang W. Platelet-to-lymphocyte ratio as a prognostic predictor of mortality for sepsis: interaction effect with disease severity-a retrospective study. BMJ Open. 2019;9(1): e022896.
Zhou Q, You X, Dong H, Lin Z, Shi Y, Su Z, Shao R, Chen C, Zhang J. Prediction of premature all-cause mortality in patients receiving peritoneal dialysis using modified artificial neural networks. Aging. 2021;13(10):14170–84.
Elhag AA, Aloafi TA, Jawa TM, Sayed-Ahmed N, Bayones FS, Bouslimi J. Artificial neural networks and statistical models for optimization studying COVID-19. Results Phys. 2021;25: 104274.
Kasai H, Ziv NE, Okazaki H, Yagishita S, Toyoizumi T. Spine dynamics in the brain, mental disorders and artificial neural networks. Nat Rev Neurosci. 2021;22(7):407–22.
Tey SF, Liu CF, Chien TW, Hsu CW, Chan KC, Chen CJ, Cheng TJ, Wu WS. Predicting the 14-Day hospital readmission of patients with pneumonia using Artificial neural networks (ANN). Int J Environ Res Public Health. 2021;18(10):5110.
Xuyi W, Seow H, Sutradhar R. Artificial neural networks for simultaneously predicting the risk of multiple co-occurring symptoms among patients with cancer. Cancer Med. 2021;10(3):989–98.
Braga MB, Fernandes RDS, Souza GN Jr, Rocha J, Dolacio CJF, Tavares IDS Jr, Pinheiro RR, Noronha FN, Rodrigues LLS, Ramos RTJ, et al. Artificial neural networks for short-term forecasting of cases, deaths, and hospital beds occupancy in the COVID-19 pandemic at the Brazilian Amazon. PLoS ONE. 2021;16(3): e0248161.
Johnson A, Pollard T, Shen L, Lehman L, Feng M, Ghassemi M, Moody B, Szolovits P, Celi L, Mark R. MIMIC-III, a freely accessible critical care database. Scientific data. 2016;3: 160035.
Chiu WT, Chung CC, Huang CH, Chien YS, Hsu CH, Wu CH, Wang CH, Chiu HW, Chan L. Predicting the survivals and favorable neurologic outcomes after targeted temperature management by artificial neural networks. J Formos Med Assoc. 2021;121(2):490–9.
Ding N, Guo C, Li C, Zhou Y, Chai X. An artificial neural networks model for early predicting in-hospital mortality in acute pancreatitis in MIMIC-III. Biomed Res Int. 2021;2021:6638919.
Zhou R, Yin W, Li W, Wang Y, Lu J, Li Z, Hu X. Prediction model for infectious disease health literacy based on synthetic minority oversampling technique algorithm. Comput Math Methods Med. 2022. https://doi.org/10.1155/2022/8498159.
Aronsson L, Andersson R, Ansari D. Artificial neural networks versus LASSO regression for the prediction of long-term survival after surgery for invasive IPMN of the pancreas. PLoS ONE. 2021;16(3): e0249206.
Lee K, Dong J, Jeong S, Chae M, Lee B, Kim H, Ko S, Song Y. Early detection of bacteraemia using ten clinical variables with an Artificial neural network approach. J Clin Med. 2019. https://doi.org/10.3390/jcm8101592.
Waxman J, Graupe D, Carley D. Automated prediction of apnea and hypopnea, using a LAMSTAR artificial neural network. Am J Respir Crit Care Med. 2010;181(7):727–33.
Cakir E, Turan I. Lactate/albumin ratio is more effective than lactate or albumin alone in predicting clinical outcomes in intensive care patients with sepsis. Scand J Clin Lab Invest. 2021;81(3):225–9.
Benediktsson S, Frigyesi A, Kander T. Routine coagulation tests on ICU admission are associated with mortality in sepsis: an observational study. Acta Anaesthesiol Scand. 2017;61(7):790–6.
Li Y, She Y, Fu L, Zhou R, Xiang W, Luo L. Association between red cell distribution width and hospital mortality in patients with sepsis. J Int Med Res. 2021;49(4):3000605211004221.
Lu J, Xun Y, Yu X, Liu Z, Cui L, Zhang J, Li C, Wang S. Albumin-globulin ratio: a novel predictor of sepsis after flexible ureteroscopy in patients with solitary proximal ureteral stones. Transl Androl Urol. 2020;9(5):1980–9.
Yin M, Si L, Qin W, Li C, Zhang J, Yang H, Han H, Zhang F, Ding S, Zhou M, et al. Predictive value of serum albumin level for the prognosis of severe sepsis without Exogenous Human Albumin Administration: a prospective cohort study. J Intensive Care Med. 2018;33(12):687–94.
Takegawa R, Kabata D, Shimizu K, Hisano S, Ogura H, Shintani A, Shimazu T. Serum albumin as a risk factor for death in patients with prolonged sepsis: an observational study. J Crit Care. 2019;51:139–44.
Kendall H, Abreu E, Cheng AL. Serum albumin trend is a predictor of mortality in ICU patients with sepsis. Biol Res Nurs. 2019;21(3):237–44.
Ishizuka M, Terasaki A, Kubota K. Exacerbation of prothrombin time-international normalized ratio before second polymyxin B cartridge hemoperfusion predicts poor outcome of patients with severe sepsis and/or septic shock. J Surg Res. 2016;200(1):308–14.
Yu J, Wang Y, Lin S, Jiang L, Sang L, Zheng X, Zhong M. Severe COVID-19 has a distinct phenotype from bacterial sepsis: a retrospective cohort study in deceased patients. Ann Transl Med. 2021;9(13):1054.
Engelmann B, Massberg S. Thrombosis as an intravascular effector of innate immunity. Nat Rev Immunol. 2013;13(1):34–45.
Atik D, Kaya H. Evaluation of the relationship of MPV, RDW and PVI parameters with disease severity in Covid-19 patients. Acta Clin Croat. 2021;60(1):103–14.
Alparslan Bekir S, Tuncay E, Gungor S, Yalcinsoy M, Sogukpinar Ö, Gundogus B, Aksoy E, Agca M, Agca Altunbey S, Turker H, et al. Can red blood cell distribution width (RDW) level predict the severity of acute exacerbation of chronic obstructive pulmonary disease (AECOPD) ? Int J Clin Pract. 2021. https://doi.org/10.20471/acc.2021.60.01.15.
Pinna A, Carlino P, Serra R, Boscia F, Dore S, Carru C, Zinellu A. Red cell distribution width (RDW) and complete blood cell count-derived measures in non-arteritic anterior ischemic optic neuropathy. Int J Med Sci. 2021;18(10):2239–44.
Cetin S, Yildiz S, Keskin K, Sigirci S, Bayraktar A, Sahin I. RDW value may increase the diagnostic accuracy of MPS. Sisli Etfal Hastanesi tip bulteni. 2021;55(1):76–80.
Jo YH, Kim K, Lee JH, Kang C, Kim T, Park HM, Kang KW, Kim J, Rhee JE. Red cell distribution width is a prognostic factor in severe sepsis and septic shock. Am J Emerg Med. 2013;31(3):545–8.
Ozdogan HK, Karateke F, Ozyazici S, Ozdogan M, Ozaltun P, Kuvvetli A, Gokler C, Ersoy Z. The predictive value of red cell distribution width levels on mortality in intensive care patients with community-acquired intra-abdominal sepsis. Ulus Travma Acil Cerrahi Derg. 2015;21(5):352–7.
Wang H, Huang J, Liao W, Xu J, He Z, Liu Y, He Z, Chen C. Prognostic value of the red cell distribution width in patients with sepsis-induced acute respiratory distress syndrome: a retrospective cohort study. Dis Markers. 2021;2021:5543822.
Villar J, Short J, Lighthall G. Lactate predicts both short- and long-term mortality in patients with and without sepsis. Infect Dis. 2019;12:1178633719862776.
Lopez R, Perez-Araos R, Baus F, Moscoso C, Salazar A, Graf J, Montes JM, Samtani S. Outcomes of sepsis and septic shock in cancer patients: focus on lactate. Front Med (Lausanne). 2021;8: 603275.
Hargreaves DS, de Carvalho JLJ, Smith L, Picton G, Venn R, Hodgson LE. Persistently elevated early warning scores and lactate identifies patients at high risk of mortality in suspected sepsis. Eur J Emerg Med. 2020;27(2):125–31.
Yang K, Fan M, Wang X, Xu J, Wang Y, Tu F, Gill PS, Ha T, Liu L, Williams DL, et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 2021. https://doi.org/10.1038/s41418-021-00841-9.
Acknowledgements
None.
Funding
None.
Author information
Authors and Affiliations
Contributions
Conception and design: YS, ND; Administrative support: CL, ND. Provision of study materials or patients: CG, SZ; Collection and assembly of data: CG, YS. Data analysis and interpretation: CG, ND. Manuscript writing: ND. Final approval of manuscript: All authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was conducted in accordance with Declaration of Helsinki 2002. MIMIC-III was an anonymized public database. To apply for access to the database, we passed the Protecting Human Research Participants exam (No.32900964). The project was approved by the institutional review boards of the Massachusetts Institute of Technology (MIT) and Beth Israel Deaconess Medical Center (BIDMC) and was given a waiver of informed consent. All the experiment protocol for involving humans was in accordance to guidelines of national/international/institutional or Declaration of Helsinki in the manuscript.
Consent for publication
Not applicable.
Competing interests
None.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Su, Y., Guo, C., Zhou, S. et al. Early predicting 30-day mortality in sepsis in MIMIC-III by an artificial neural networks model. Eur J Med Res 27, 294 (2022). https://doi.org/10.1186/s40001-022-00925-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-022-00925-3