Abstract
Background
Aberrant DNA methylation plays a crucial role in the progression of myeloid neoplasms. Previously, our literature reported that slit guidance ligand 2 (SLIT2) promoter methylation was associated with disease progression and indicated a poor prognosis in patients with myelodysplastic syndrome. Herein, we further investigated the clinical implications and role of SLIT2 promoter methylation in patients with chronic myeloid leukemia (CML).
Methods
The level of SLIT2 promoter methylation was determined in 104 CML patients, and its clinical significance was analyzed. Moreover, demethylation studies were performed in K562 cells to determine the epigenetic mechanism by which SLIT2 promoter methylation is regulated in CML.
Results
The level of SLIT2 promoter methylation was similar between CML patients and controls. However, deeper analysis revealed that the SLIT2 promoter methylation level in the accelerated phase (AP) and blast crisis (BC) was markedly higher than that in the chronic phase (CP) and controls. Additionally, a marked difference was identified between the SLIT2 promoter hypermethylated and non-hypermethylated groups among CML patients grouped by clinical stage. The frequency of SLIT2 hypermethylation was markedly increased with the progression of clinical stage, that is, it was the lowest in CP samples (12/80, 15%), higher in AP samples (4/8, 50%) and the highest in BC samples (11/16, 69%). Importantly, the level/density of SLIT2 promoter methylation was significantly higher in the advanced stage than in the early stage among the 6 tested paired CML patients. Epigenetically, the expression of the SLIT2-embedded non-coding genes SLIT2-IT1 and miR-218 expression was decreased in patients with CML. SLIT2 promoter hypermethylated cases had a markedly lower SLIT2-IT1 expression level than SLIT2 promoter non-hypermethylated cases. Moreover, SLIT2-IT1 and miR-218 expression was remarkably upregulated in a dose-dependent manner after demethylation treatment of K562 cells.
Conclusions
Hypermethylation of the SLIT2 promoter is correlated with disease progression in CML. Furthermore, SLIT2 promoter methylation may function by regulating the expression of the SLIT2-embedded non-coding genes SLIT2-IT1 and miR-218 during CML progression.
Similar content being viewed by others
Background
Chronic myeloid leukemia (CML) is initiated by the reciprocal translocation t(9;22)/Philadelphia (Ph) chromosome, which leads to the formation of the BCR::ABL1 fusion protein with aberrant tyrosine kinase activity [1]. The treatment of CML is also based on the inhibition of aberrant tyrosine kinase activity as targeted therapy [1]. The typical clinical course of CML includes the initial stage of the chronic phase (CP) and the advanced/aggressive stage of the accelerated phase (AP) and blast crisis (BC) during disease progression [1]. Although CML is cytogenetically/genetically homogenous at the earlier stage, considerable genetic and/or epigenetic heterogeneity is identified in the later stage of CML [1, 2]. Cytogenetic and genetic abnormalities are pathogenetically associated with the progression of CML [2, 3]. Recently, aberrant DNA methylation, which plays a crucial role in the progression of CML, has attracted our attention [4, 5].
The slit guidance ligand (SLIT) family members (SLIT1/SLIT2/SLIT3) are highly conserved secreted glycoproteins that regulate various physiologic processes, such as neuronal axon guidance, cell proliferation, cell migration, and vascularization, by binding to roundabout (ROBO) receptors (ROBO1/ROBO2/ROBO3/ROBO4) [6]. The SLIT/ROBO signaling pathway was originally recognized in the nervous system and functions in neuronal axon guidance and is also considered an important regulator of multiple physiological and oncogenic processes [6, 7]. Recently, an increasing number of studies have reported the dysregulation of SLIT/ROBO signaling pathways in a variety of human cancers [7]. Epigenetic silencing of SLITs mediated by promoter hypermethylation plays a vital role in cancer initiation and progression [10]. Accordingly, a number of studies have shown that SLITs/ROBOs are frequently downregulated and have anticancer roles in the advanced stage of several solid tumors [8, 9]. However, several other studies have demonstrated an oncogenic role during cancer development [8, 9]. Interestingly, the SLIT2-embedded non-coding RNA (ncRNA) miR-218 was found to be downregulated and to act as a tumor suppressor gene in human cancers in most studies [11]. In addition, the other SLIT2-embedded ncRNA SLIT2-IT1 has rarely been investigated.
Previously, our study reported that hypermethylation of the SLIT2 promoter was associated with disease progression in myelodysplastic syndrome (MDS) and predicted poor clinical outcome in both MDS and acute myeloid leukemia (AML) [12]. Moreover, SLIT2 promoter methylation exerted its function by repressing the expression of two SLIT2-embedded ncRNAs, SLIT2-IT1 and miR-218 (SLIT2-IT1/miR-218), in MDS and AML [12]. However, the pattern and clinical implications of SLIT2 promoter methylation in CML remain poorly defined. Herein, on the basis of previous research, we further determined the pattern, clinical implication and role of SLIT2 promoter methylation in patients with CML.
Materials and methods
Subjects and samples
The current study included 104 de novo CML patients (80 in CP stage, 8 in AP stage and 16 in BC stage) and 51 healthy donors (age and sex-matched). The diagnosis and clinical stages of CML were established by clinical manifestation and laboratory examination of peripheral blood (PB)/bone marrow (BM), and were confirmed by molecular detection of the BCR::ABL1 transcript. The BCR::ABL transcript detection was quantified using real-time quantitative PCR (RT-qPCR) established previously [13]. BM samples collected from the subjects were further used for the extraction of BM mononuclear cells (BMMNCs) using Lymphocyte Separation Medium (Solarbio, Beijing, China) by gradient centrifugation.
Cell line, cell culture and demethylation treatment
The human CML cell line K562 was cultured in RPMI 1640 medium (Solarbio, Beijing, China) with 10% fetal calf serum (ExCell, Shanghai, China) and grown in a 5% CO2 humidified atmosphere at 37 °C. For demethylation treatment, K562 cells were treated with 5-aza-2’-deoxycytidine (5-aza-dC) (Sigma‒Aldrich, St. Louis, MO) at final concentrations of 0 μM, 1 μM, 2 μM, and 4 μM for 3 days. All treated cells were cultured until harvested for extraction of total RNA and DNA.
RNA isolation, reverse transcription and RT-qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA), followed by reverse transcription to synthesize cDNA for miRNA and long non-coding RNA (lncRNA) detection [12, 14]. RT-qPCR was performed to examine SLIT2-IT1/miR-218 expression by AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ). The primers for SLIT2-IT1/miR-218 expression were previously reported [12]. Relative SLIT2-IT1/miR-218 transcript expression was calculated using the 2− ∆∆CT formula according to the ABL1 transcript.
DNA isolation, chemical modification and RT-qMSP
The isolation and modification of genomic DNA was performed using Puregene Blood Core Kit B and EpiTect Bisulfite Kit (QIAGEN, Duesseldorf, Germany) as described previously [15, 16]. Real-time quantitative methylation-specific PCR (RT-qMSP) was first used to evaluate SLIT2 promoter methylation with AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ). The primers for SLIT2 promoter methylation detection were as reported [12]. Relative SLIT2 promoter methylation was counted using the 2− ∆∆CT formula as referred to ALU methylation.
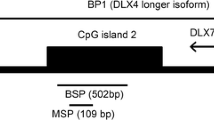
BSP
Bisulfite sequencing PCR (BSP) was further performed to detect SLIT2 promoter methylation using TaKaRa Taq™ Hot Start Version (Tokyo, Japan). The primers for SLIT2 promoter methylation detected by BSP were reported previously [17]. The details of BSP can be found in our previous study [17]. Six independent clones from each specimen were selected for Sanger sequencing (BGI, Shanghai, China).
Statistics
Statistics were accomplished using SPSS 20.0 and GraphPad Prism 5.0 software packages. The differences in continuous variables between the two groups were compared by Mann‒Whitney’s U test. The differences in categorical variables between the two groups were compared by Pearson Chi-square analysis or Fisher’s exact test. The association of SLIT2 promoter methylation with SLIT2-IT1/miR-218 expression was analyzed by Spearman correlation test. Among all statistical analyses, a two-tailed P value < 0.05 was considered statistically significant.
Results
SLIT2 promoter methylation in CML patients
Previously, we reported the pattern of SLIT2 promoter methylation in patients with MDS and AML and revealed that SLIT2 promoter methylation was correlated with disease progression [12]. Herein, we further detected SLIT2 promoter methylation in CML patients by RT-qMSP as previously described. The results showed that the SLIT2 promoter methylation level was similar between CML patients and controls (P = 0.187, Fig. 1). However, further analysis revealed that the SLIT2 promoter methylation level in the CML-AP and CML-BC stages was markedly higher than that in the CML-CP stage (P = 0.014 and < 0.001, respectively, Fig. 1) and in controls (P = 0.022 and < 0.001, respectively, Fig. 1). The above results indicated that SLIT2 promoter methylation is correlated with an advanced stage of CML and may correlate with disease progression.
Relative SLIT2 promoter methylation level in controls and CML patients. SLIT2 promoter methylation was detected by RT-qMSP in controls and whole CML patients as well as different stages of CML patients (CP stage, AP sage, and BC stage) were presented with scatter plots. The median level of SLIT2 promoter methylation in each group was shown with horizontal line
Association between SLIT2 promoter methylation and clinicopathological characteristics of CML patients
To determine the correlation between SLIT2 promoter methylation and clinicopathological characteristics of CML, the whole cohort of CML patients was divided into two groups based on the previously set cut-off points [12]. No statistical differences were found between the SLIT2 promoter hypermethylated and non-hypermethylated groups with respect to sex, age, hemoglobin, karyotype and BCR-ABL transcript status (Table 1). However, SLIT2 promoter hypermethylated cases exhibited lower white blood cells (WBCs) and platelets than SLIT2 promoter non-hypermethylated cases (P < 0.001 and = 0.006, respectively, Table 1). Notably, a marked difference was identified between the SLIT2 promoter hypermethylated and non-hypermethylated groups in CML patients grouped by clinical stage (P < 0.001, Table 1). The frequency of SLIT2 hypermethylation was markedly increased with the progression of clinical stage, that is, it was the lowest in CML-CP samples (12/80, 15%), higher in CML-AP samples (4/8, 50%) and the highest in CML-BC samples (11/16, 69%) (P < 0.001, Table 1). These results further confirmed that SLIT2 promoter methylation was correlated with an advanced stage of CML and may correlate with disease progression.
SLIT2 promoter methylation alteration during disease progression in paired CML patients
Given the results above, we hypothesized that SLIT2 promoter methylation was correlated with disease progression in CML. To test this hypothesis, we further examined SLIT2 promoter methylation in paired CML patients during disease progression. By RT-qMSP, the level of SLIT2 promoter methylation was significantly upregulated in the advanced stage compared with the early stage among the tested 6 paired CML patients (Fig. 2). Moreover, the SLIT2 promoter methylation density in these paired patients was further detected by BSP (Fig. 3) and was closely correlated with the results detected by RT-qMSP (R = 0.895, P < 0.001, Additional file 1: Fig S1). Taken together, these results suggest that SLIT2 promoter methylation is correlated with disease progression in CML.
SLIT2 promoter methylation density alterations during disease progression in six paired CML patients. White cycle: unmethylated CpG dinucleotide; Black cycle: methylated CpG dinucleotide. SLIT2 promoter methylation density in CP stage in Patient a-f were 17.3%, 5.3%, 9.7%, 11%, 11.3% and 9%, whereas in AP/BC stage in Patient a-f were 70.7%, 38%, 50.7%, 29.3%, 26% and 33.7% (P = 0.004, Paired T test)
Epigenetic regulatory effects of SLIT2 promoter methylation in CML
Previously, we revealed that SLIT2 promoter methylation was associated with SLIT2-embedded ncRNAs SLIT2-IT1/miR-218 expression but not SLIT2 expression in MDS and AML. Herein, we further detected SLIT2-IT1/miR-218 expression in 51 CML patients with available mRNA samples matched to DNA samples. SLIT2-IT1 expression was markedly decreased (P = 0.030, Fig. 4a), whereas miR-218 expression was nearly undetectable in CML patients. Moreover, although SLIT2-IT1 expression exhibited a weak negative association with SLIT2 promoter methylation in CML patients (R = − 0.289, P = 0.039, n = 51), cases with SLIT2 promoter hypermethylation had a markedly lower SLIT2-IT1 expression level than those without SLIT2 promoter hypermethylation (P = 0.004, Fig. 4b). To further verify the epigenetic regulatory effects of SLIT2 promoter methylation on the ncRNAs SLIT2-IT1/miR-218, we performed demethylation treatment of the CML cell line K562 with 5-aza-dC. With the decreased density of SLIT2 promoter methylation, SLIT2-IT1/miR-218 expression was markedly upregulated in a dose-dependent manner after 5-aza-dC treatment (Fig. 4c–f). Collectively, these results support the epigenetic regulatory effects of SLIT2 promoter methylation on the expression of SLIT2-embedded ncRNAs SLIT2-IT1/miR-218 in CML.
Transcriptional regulatory effects of SLIT2 promoter methylation on SLIT2-IT1/miR-218 expression in CML. a Relative SLIT2-IT1 expression level in CML patients; b relative SLIT2-IT1 expression between SLIT2 promoter non-hypermethylated and hypermethylated groups; c SLIT2-IT1 expression before and after 5-aza-dC treatment with different dose; d miR-218 expression before and after 5-aza-dC treatment with different dose; e SLIT2 promoter methylation density before 5-aza-dC treatment; f SLIT2 promoter methylation density after 5-aza-dC treatment (4 μM)
Discussion
The SLIT/ROBO signaling pathway has been implicated in the regulation of developmental processes and physiological processes [6, 7]. SLIT/ROBO signaling plays crucial roles in a number of cell signaling pathways including axon guidance, angiogenesis, cell proliferation, cell apoptosis and cell motility [6, 7]. Moreover, inactivation of SLITs/ROBOs expression mediated by promoter methylation in cells can lead to cancer initiation and progression [10]. Notably, several studies have demonstrated that SLITs/ROBOs are frequently downregulated in the advanced stage of various solid tumors [8, 9]. This evidence indicated that the SLIT/ROBO signaling pathway may play a crucial role in cancer progression than cancer initiation. Previously, we reported that SLIT2 promoter methylation through the inactivation of SLIT2-IT1/miR-218 expression may play a key role in MDS progression by affecting cell proliferation, apoptosis and colony formation both in vitro and in vivo [12]. In the current study, we observed that SLIT2 promoter hypermethylation was associated with lower WBCs and platelet in CML, which suggests that SLIT2 promoter hypermethylation may be associated with hematopoietic stem cells differentiation fate. Accordingly, further functional studies are needed to determine the direct role of aberrant SLIT2 promoter methylation in leukemogenesis during CML progression.
To date, the mechanisms involved in CML progression have been preliminarily identified. Cytogenetic aberrations, such as double t(9;22)/Ph chromosome, trisomy chromosome 8, i(17q), trisomy chromosome 19, t(3;21) and t(7;11), and molecular alterations, including TP53 mutations, RAS mutations and increased BCR::ABL1 transcript levels, are pathogenetically correlated with the progression of CML [2, 3]. Moreover, epigenetic alterations, such as aberrant DNA methylation, have also been identified to play a vital role in the disease evolution of CML [4, 5]. For instance, Li et al. revealed that SHP-1 hypermethylation was involved in CML evolution through the regulation of the BCR::ABL1, AKT, MAPK, MYC and JAK2/STAT5 signaling pathways [18]. Additionally, our research group has also revealed the correlation of SOX30, ID4 and DLX4 hypermethylation with disease progression in CML [19,20,21]. A recent study demonstrated that promoter hydroxymethylation of tumor suppressor genes DAPK1, RIZ1, P16INK4A, RASSF1A and p14ARFARF was a characteristic feature of CML disease progression and indicated poor imatinib response and poor overall survival of CML patients to imatinib therapy [22]. On the basis of our previous study [12], we further investigated SLIT2 promoter methylation in another myeloid malignancy CML. In accordance with the results in MDS [12], SLIT2 promoter methylation was also correlated with advanced clinical stage of CML, and played a crucial role in disease progression. Interestingly, Heller et al. observed up to 897 genes that were methylated at the time of progression but not at the time of diagnosis in CP-CML patients who progressed to AP/BC-CML using next-generation sequencing [4]. However, SLIT2 promoter hypermethylation was not identified in this study [4], which may be attributed to differences in ethical considerations. Since this is the first report of SLIT2 promoter hypermethylation in CML progression, prospective investigations are needed to confirm and expand our results.
DNA hypermethylation mainly functions by inactivating gene expression in cancer development. Although a few investigations have demonstrated the association between SLIT2 promoter methylation and SLIT2 expression in some types of solid tumors [23], our recent study revealed that SLIT2 promoter methylation was correlated with the expression of SLIT2-embedded ncRNAs SLIT2-IT1/miR-218 but not with SLIT2 in AML [12]. Herein, we also explored the expression of SLIT2-IT1 and miR-218 expression in CML. The results showed that SLIT2-IT1/miR-218 was significantly decreased in CML patients, and was negatively correlated with SLIT2 promoter methylation. Moreover, demethylation studies also confirmed the epigenetic mechanism of SLIT2 promoter methylation in regulating ncRNAs SLIT2-IT1/miR-218 expression in CML. Taken together, these results indicated that SLIT2 promoter hypermethylation may function by repressing SLIT2-IT1/miR-218 expression during CML progression.
Conclusion
Hypermethylation of the SLIT2 promoter is correlated with disease progression in CML. Furthermore, SLIT2 promoter methylation may regulate the expression of SLIT2-embedded non-coding genes SLIT2-IT1/miR-218 during CML progression.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CML:
-
Chronic myeloid leukemia
- CP:
-
Chronic phase
- AP:
-
Accelerated phase
- BC:
-
Blast crisis
- Ph:
-
Philadelphia
- MDS:
-
Myelodysplastic syndrome
- AML:
-
Acute myeloid leukemia
- SLIT:
-
Slit guidance ligand
- ROBO:
-
Roundabout
- ncRNAs:
-
Non-coding RNAs
- PB:
-
Peripheral blood
- BM:
-
Bone marrow
- BMMNCs:
-
BM mononuclear cells
- 5-aza-dC:
-
5-Aza-2’-deoxycytidine
- lncRNA:
-
Long non-coding RNA
- RT-qPCR:
-
Real-time quantitative PCR
- RT-qMSP:
-
Real-time quantitative methylation-specific PCR
- BSP:
-
Bisulfite sequencing PCR
- WBC:
-
White blood cell
References
Cortes J, Pavlovsky C, Saußele S. Chronic myeloid leukaemia. Lancet. 2021;398(10314):1914–26.
Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103(11):4010–22.
Bavaro L, Martelli M, Cavo M, Soverini S. Mechanisms of disease progression and resistance to tyrosine kinase inhibitor therapy in chronic myeloid leukemia: an update. Int J Mol Sci. 2019;20(24):6141.
Heller G, Topakian T, Altenberger C, Cerny-Reiterer S, Herndlhofer S, Ziegler B, Datlinger P, Byrgazov K, Bock C, Mannhalter C, Hörmann G, Sperr WR, Lion T, Zielinski CC, Valent P, Zöchbauer-Müller S. Next-generation sequencing identifies major DNA methylation changes during progression of Ph+ chronic myeloid leukemia. Leukemia. 2016;30(9):1861–8.
Koschmieder S, Vetrie D. Epigenetic dysregulation in chronic myeloid leukaemia: a myriad of mechanisms and therapeutic options. Semin Cancer Biol. 2018;51:180–97.
Blockus H, Chédotal A. Slit-robo signaling. Development. 2016;143(17):3037–44.
Tong M, Jun T, Nie Y, Hao J, Fan D. The role of the slit/robo signaling pathway. J Cancer. 2019;10(12):2694–705.
Gara RK, Kumari S, Ganju A, Yallapu MM, Jaggi M, Chauhan SC. Slit/Robo pathway: a promising therapeutic target for cancer. Drug Discov Today. 2015;20(1):156–64.
Jiang Z, Liang G, Xiao Y, Qin T, Chen X, Wu E, Ma Q, Wang Z. Targeting the SLIT/ROBO pathway in tumor progression: molecular mechanisms and therapeutic perspectives. Ther Adv Med Oncol. 2019;11:1758835919855238.
Narayan G, Goparaju C, Arias-Pulido H, Kaufmann AM, Schneider A, Dürst M, Mansukhani M, Pothuri B, Murty VV. Promoter hypermethylation-mediated inactivation of multiple slit-robo pathway genes in cervical cancer progression. Mol Cancer. 2006;5:16.
Lu YF, Zhang L, Waye MM, Fu WM, Zhang JF. MiR-218 mediates tumorigenesis and metastasis: perspectives and implications. Exp Cell Res. 2015;334(1):173–82.
Zhang TJ, Xu ZJ, Wen XM, Gu Y, Ma JC, Yuan Q, Lin J, Zhou JD, Qian J. SLIT2 promoter hypermethylation-mediated SLIT2-IT1/miR-218 repression drives leukemogenesis and predicts adverse prognosis in myelodysplastic neoplasm. Leukemia. 2022;36(20):2488–98.
Qian J, Wang YL, Lin J, Yao DM, Xu WR, Wu CY. Aberrant methylation of the death-associated protein kinase 1 (DAPK1) CpG island in chronic myeloid leukemia. Eur J Haematol. 2009;82(2):119–23.
Zhou JD, Zhang TJ, Xu ZJ, Deng ZQ, Gu Y, Ma JC, Wen XM, Leng JY, Lin J, Chen SN, Qian J. Genome-wide methylation sequencing identifies progression-related epigenetic drivers in myelodysplastic syndromes. Cell Death Dis. 2020;11(11):997.
Zhang TJ, Xu ZJ, Gu Y, Wen XM, Ma JC, Zhang W, Deng ZQ, Leng JY, Qian J, Lin J, Zhou JD. Identification and validation of prognosis-related DLX5 methylation as an epigenetic driver in myeloid neoplasms. Clin Transl Med. 2020;10(2): e29.
Zhang TJ, Xu ZJ, Gu Y, Ma JC, Wen XM, Zhang W, Deng ZQ, Qian J, Lin J, Zhou JD. Identification and validation of obesity-related gene LEP methylation as a prognostic indicator in patients with acute myeloid leukemia. Clin Epigenetics. 2021;13(1):16.
Zhou JD, Wang YX, Zhang TJ, Li XX, Gu Y, Zhang W, Ma JC, Lin J, Qian J. Identification and validation of SRY-box containing gene family member SOX30 methylation as a prognostic and predictive biomarker in myeloid malignancies. Clin Epigenetics. 2018;10:92.
Li Y, Yang L, Pan Y, Yang J, Shang Y, Luo J. Methylation and decreased expression of SHP-1 are related to disease progression in chronic myelogenous leukemia. Oncol Rep. 2014;31(5):2438–46.
Zhang TJ, Wen XM, Zhou JD, Gu Y, Xu ZJ, Guo H, Ma JC, Yuan Q, Chen Q, Lin J, Qian J. SOX30 methylation correlates with disease progression in patients with chronic myeloid leukemia. Onco Targets Ther. 2019;12:4789–94.
Zhou JD, Zhang TJ, Li XX, Ma JC, Guo H, Wen XM, Zhang W, Yang L, Yan Y, Lin J, Qian J. Epigenetic dysregulation of ID4 predicts disease progression and treatment outcome in myeloid malignancies. J Cell Mol Med. 2017;21(8):1468–81.
Zhou JD, Wang YX, Zhang TJ, Yang DQ, Yao DM, Guo H, Yang L, Ma JC, Wen XM, Yang J, Lin J, Qian J. Epigenetic inactivation of DLX4 is associated with disease progression in chronic myeloid leukemia. Biochem Biophys Res Commun. 2015;463(4):1250–6.
Guru SA, Sumi MP, Mir R, Beg MMA, Koner BC, Saxena A. Aberrant hydroxymethylation in promoter CpG regions of genes related to the cell cycle and apoptosis characterizes advanced chronic myeloid leukemia disease, poor imatinib respondents and poor survival. BMC Cancer. 2022;22(1):405.
Mohamed G, Talima S, Li L, Wei W, Rudzki Z, Allam RM, Simmons W, Tao Q, Murray PG. Low expression and promoter hypermethylation of the tumour suppressor SLIT2, are associated with adverse patient outcomes in diffuse large B cell lymphoma. Pathol Oncol Res. 2019;25(3):1223–31.
Acknowledgements
None.
Funding
The work was supported by National Natural Science Foundation of China (81900166, 81900163, 81970118, 82270179), Natural Science Foundation of Jiangsu Province (BK20221287), Research Project of Jiangsu Commission of Health (M2022123), Zhenjiang Clinical Research Center of Hematology (SS2018009), Medical Field of Zhenjiang “Jin Shan Ying Cai” Project, Medical Education Collaborative Innovation Fund of Jiangsu University (JDY2022011).
Author information
Authors and Affiliations
Contributions
JZ and JQ conceived and designed the experiments; DW, MC and TZ performed the experiments; YW and ZX analyzed the data and provided bioinformatics analysis; QY collected the clinical data; JM and JL provided the technical and financial supports; JZ wrote and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical approval and consent to participate
The present study approved by the Ethics Committee of the Affiliated People’s Hospital of Jiangsu University. Written informed consents were obtained from all enrolled individuals prior to their participation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Correlation between SLIT2 methylation density detected by BSP and SLIT2 methylation level detected by RT-qMSP.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wu, Dl., Wang, Y., Zhang, Tj. et al. SLIT2 promoter hypermethylation predicts disease progression in chronic myeloid leukemia. Eur J Med Res 27, 259 (2022). https://doi.org/10.1186/s40001-022-00899-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-022-00899-2