Abstract
Soybean has been recognized as a useful platform for heterologous protein production. This study compared the pollen characteristics of transgenic and non-transgenic soybean and investigated the rate of gene flow from transgenic soybean events, developed to obtain recombinant proteins (such as human epidermal growth factor, insulin-like growth factor 1, or thioredoxin) for use in the skin care industry, to non-transgenic soybean under field conditions, and determined the distance at which gene flow could occur. The lack of significant differences in pollen grain size, viability and pollen germination rates between transgenic and non-transgenic cultivars indicates that the overexpression of transgenes did not alter pollen characteristics in soybean. The highest rates of gene flow from the three transgenic soybean events to non-transgenic soybean ranged from 0.22 to 0.46% at the closest distance (0.5 m). Gene flow was observed up to 13.1 m from the transgenic plots. Our data fell within the ranges reported in the literature and indicate that an isolation distance greater than at least 13 m from transgenic soybean is required to prevent within-crop gene flow in soybean. As the potential markets for transgenic crops as a recombinant protein factory increase, gene flow from transgenic to non-transgenic conventional crops will become a key decision factor for policy makers during the approval process of transgenic crops. Our study may provide useful baseline data for the prevention of transgenic soybean seed contamination caused by transgene flow.
Similar content being viewed by others
Introduction
Soybean (Glycine max (L.) Merr.) is a predominantly self-pollinated crop, and pollination occurs before the flowers open. The rates of natural cross-pollination between closely placed soybean cultivars are generally found to be less than 1% under field conditions [1,2,3,4,5]. However, maximum rates of natural cross-pollination as high as 4.52% [6] and 6.32% [7] have also been reported. Moreover, much higher rates were observed in male sterile soybean lines [8, 9]. Because the airborne release of soybean pollen is very limited, soybean pollination is not mediated by wind [10]. Insect pollinators, particularly many bee species, play important roles in soybean pollination [11,12,13,14,15]. The presence of adequate insect pollinators, distance from pollen source, location, season, pollen recipient genotype, and ambient temperature can affect the natural cross-pollination of soybean [8, 16,17,18].
Since the commercial cultivation of glyphosate-resistant transgenic soybean (GTS 40-3-2 event), the adoption of transgenic soybean has greatly increased worldwide and reached 95.9 million hectares in 2018 [19]. Although many benefits of transgenic crop cultivation have been recognized, considerable economic concerns regarding transgenic seed contamination have been raised, and coexistence of non-transgenic (conventional or organic) and transgenic cropping system has become an important issue in countries that cultivate transgenic crops. Current studies on gene flow from transgenic to non-transgenic crop cultivars aim to gain information on how to prevent inadvertent admixture of those crops through outcrossing. A few studies have reported gene flow from transgenic soybean to non-transgenic soybean cultivars in the fields of Brazil [20,21,22,23], China [24,25,26,27,28], Japan [29], and Korea [30, 31]. These studies mostly used herbicide-resistant transgenic soybean as pollen donors and measured the rates of gene flow and the distance at which gene flow can occur.
Using seeds as bioreactors for the production of recombinant proteins has been explored [32,33,34]. Among the whole plant expression systems, seed-based expression platforms have an advantage over leaf-based platforms. The plant seed has a high protein content but low protease activity; therefore, protection from proteolytic degradation enables much longer storage of recombinant protein than that with the protein produced in leaf tissues [35]. Soybean is currently recognized as a useful platform to produce heterologous proteins [36, 37].
Three transgenic soybean events expressing human epidermal growth factor (EGF), insulin-like growth factor 1 (IGF-1), or thioredoxin (TRX) have been developed to obtain recombinant proteins for use in the skin care industry. In these events, the transgenes are controlled by the β-conglycinin promoter for seed-specific expression. Moreover, EGF stimulates the growth of skin and corneal epithelium and acts as a potent mitogenic factor [38, 39]. In addition, IGF-1 protein has insulin-like effects, such as the stimulation of glucose consumption in adipose tissue, and is involved in mediating growth and development [40, 41]. TRX protein is involved in many redox reactions and has antioxidant, anti-inflammatory, and anti-allergic properties [42, 43].
Before commercializing transgenic soybean events, their potential risks to human health and the environment should be thoroughly assessed. Recently, the effects of such transgenic events on arthropod communities [44] as well as the invasiveness [45] of these events have been reported. In addition, measures to prevent the entry of such events into the food or feed supply chain are needed. In this study, we aimed to investigate the rate of gene flow from transgenic soybean events to non-transgenic soybean and to determine the distance at which gene flow can occur. In addition, we investigated pollen characteristics including size, viability, and germination rates, which are important characteristics related to pollen dispersal and reproductive success.
Materials and methods
Plant materials
Using a Korean soybean cultivar ‘Kwangan’, transgenic soybean (Glycine max (L.) Merr.) events were developed through Agrobacterium tumefaciens-mediated transformation. The non-transgenic host cultivar ‘Kwangan’ belongs to maturity group VI, with days to maturity ranging from 156 to 163 days [46]. The transgenic soybean events contained either the human epidermal growth factor (egf) gene (event CT-1001), human insulin-like growth factor 1 (igf-1) gene (event CT-2062), or human thioredoxin (trx) gene (event CT-4025) gene under the control of the β-conglycinin promoter for seed-specific protein expression and the PinII terminator. All transgenic events contained the bar gene for phosphinothricin selection with the cauliflower mosaic virus 35S promoter and nos terminator. We obtained seeds of transgenic soybean events and the non-transgenic host cultivar, ‘Kwangan’, from the Research & Development Center, CELLTRION, in March 2017.
Pollen characteristics
Transgenic and non-transgenic soybean seeds were sterilized and sown on seed trays filled with commercial potting soil. 2 weeks later, seedlings were transplanted in 7-L pots with five replicates and grown in a greenhouse. During the flowering stage, two flowers per plant were randomly collected between 09:00 and 10:00 a.m. and air-dried for 2 h before the subsequent measurements. The pollen characteristics of CT-1001, CT-4024, and CT-2062 soybeans were compared with those of ‘Kwangan’ in June, July, and September 2016, respectively. Because it was difficult to handle all events in a single day and as we did not aim to compare pollen characteristics among transgenic events; we compared the non-transgenic soybean and one transgenic soybean event at a time.
Pollen was added on a glass slide covered with Vaseline. The slides were observed under a microscope (Olympus BX41, Japan) and photographs were taken using a digital camera. The polar axis length and equatorial diameter of 20 pollen grains per plant were measured using an image analysis program (DIXI eXcope X3, Korea). The diameter of the pollen grains was calculated as the mean value of the polar axes and equatorial diameters.
Pollen grains were stained with Lugol’s solution (I2/KI) to determine pollen viability. Pollen wase dusted onto a glass slide on which 50 μl of Lugol’s solution was added. After 30 min of staining, the glass slide was covered with a cover glass. In total, 100 pollen grains per plant were observed under a microscope (Olympus BX41, Japan), and the percentage of viable pollen was calculated. Pollen grains that were stained black were judged as viable and those stained bright yellow were judged as dead pollen.
To estimate the germination rate of pollen, we used the medium [47] containing 15 g sucrose, 0.03 g Ca(NO3)2·4H2O, and 0.01 g H3BO3 in 100 mL deionized water. To this medium, 0.5 g of agar was added and heated until the agar was completely dissolved. The medium (7 mL) was poured into a Petri dish (4 cm diameter) and allowed to cool to room temperature. Pollen was dusted on the germination medium and the plates were incubated at 30 °C for 24 h in an incubator. After 24 h, 100 pollen grains per plant were observed under a microscope, and the percentage of germinated pollen grains was calculated. Pollen grains were considered germinated if their tube lengths were greater than or equal to the grain diameter.
Field experiments
A field experiment was conducted in an experimental field at the Korea Research Institute of Bioscience & Biotechnology (KRIBB), Cheongju, Republic of Korea (36° 43′ 04″ N, 127° 26′ 07″ E, altitude: 37 m) in 2017 to investigate the rate of gene flow from transgenic soybean events to non-transgenic soybean under field conditions. During the growing season (June–October), the total rainfall was 1133 mm and the monthly mean air temperature ranged from 15.9 to 27.1 °C [48].
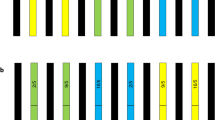
In May 2017, we established three 4 × 4 m central plots and six 10.5 × 4 m plots with five rows mulched with black plastic films (Fig. 1). On May 30, 2017, transgenic and non-transgenic soybean seeds were sown on seed trays filled with commercial potting soil (BioPlus, Hungnong, Korea) and grown in a glasshouse. Soybean seedlings were transplanted in the field on June 13, 2017. In each central plot, 105 transgenic soybean seedlings were transplanted at 20-cm spacing. In the surrounding north-east and south-west plots, 200 non-transgenic soybeans were transplanted at 50-cm spacing in five rows from the edge of the central transgenic soybean plot. Both transgenic and non-transgenic soybeans were cultivated according to conventional practices. Insecticides were sprayed to control stink bugs (Pentatomidae) in July–September. The dates of flowering initiation, 50% flowering, and flowering termination of the transgenic and non-transgenic soybean plants cultivated in the middle rows for each plot were investigated in July and August. Soybean pods of the non-transgenic recipients were collected at various distances (0.5–10.5 m) from the transgenic soybean plots on November 1, 2017. Then, soybean seeds were individually harvested from the pods by hand and stored until analysis.
Experimental design of the field trial conducted in 2017. Three transgenic soybean events (CT-1001, CT-2062, and CT-4025) were planted in each central plot. Non-transgenic cultivar ‘Kwangan’ (WT) were planted in the surrounding north-east (NE) and south-west (SW) plots. A red line indicates the row at which the trx gene of the CT-4025 event was found in the hybrid progenies
Hybrid detection
Harvested seeds from non-transgenic soybean plots were screened to determine hybrid progenies between transgenic and non-transgenic soybeans. In a greenhouse, 200 seeds were placed in a plastic tray (32 × 48 × 8 cm) filled with commercial potting soil (BioPlus, Hungnong, Korea) and watered daily. When seedlings reached the third trifoliate (V3) stage, glufosinate (Basta; Bayer Crop Science, Korea) was applied at a rate of 3 mL L−1 using a garden sprayer. The surviving seedlings were considered to be resistant to glufosinate. To estimate the number of evaluated progenies, seed germination rates were measured.
The presence of transgenes was confirmed by PCR. Primers were designed to detect transgenes (egf, igf-1, and trx) and another set of primers was used to detect the lectin gene as an internal PCR-positive control (Table 1). All primers were synthesized by Bioneer (Daejeon, Korea). Leaf samples were collected from the glufosinate-resistant soybean seedlings and DNA was extracted using a FastDNA kit (MP Bio., USA). PCR was run with a final volume of 50 μL containing 2 μL of gDNA, 25 μL of AccuPower PCR Master Mix (Bioneer, Korea), 19 µL of distilled water (Bioneer, Korea), and 2 µL of 10 pmol for each primer. The initial touchdown cycle comprised initial denaturation at 95 °C for 3 min, denaturation at 95 °C for 30 s, annealing at 58 °C (for lectin) or 59 °C (for transgenes) for 30 s, and extension at 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. As the positive control and the negative control, transgenic soybean and distilled water were used, respectively. We calculated the hybridization rates as the percentages of hybrids per number of evaluated progenies.
Statistical analyses
Data on size, viability, and germination rate of pollen were analyzed using the Real Statistics Resource Pack Software (Release 6.2.2) [49]. Data were assessed for normality and homoscedasticity using the Shapiro–Wilk and Levene’s tests, respectively. The overall effect of soybean genotype on the size, viability, and germination rate of pollen was evaluated by using the Student’s t-test. Non-normally distributed data were evaluated using the Mann–Whitney test.
Results
Pollen characteristics
No significant differences were found in pollen size, viability, and germination rates between non-transgenic and transgenic soybeans (Table 2). The mean pollen grain diameter of non-transgenic soybean ranged from 24.8 (June) to 27.3 (September) μm, and that of transgenic soybean events ranged from 25.1 (CT-1001) to 26.6 (CT-4025) μm. Pollen viability determined by staining was 95.2 to 97.6% for non-transgenic soybean and 95.0 to 97.4% for transgenic soybean events. Pollen germination rates of non-transgenic and transgenic soybean ranged from 86.2 to 96.0% and 87.6 to 96.4%, respectively.
Gene flow rates
The non-transgenic and three transgenic soybean events flowered from July 21, 2017 and July 23, 2017, respectively. The 50% flowering date of non-transgenic soybean and CT-1001 transgenic soybean was July 23, 2017, and that of CT-2062 and CT-4025 transgenic soybean was July 24, 2017 (Table 3).
From the seeds collected from non-transgenic soybean planted with CT-1001, 47,035 progenies were assessed to identify hybrids (Table 4). PCR analysis showed the presence of a 143-bp egf gene fragment in 15 samples, and the overall gene flow rate was 0.032%. The highest gene flow rate was observed for soybean planted at a distance of 0.5 m (0.210%) from the transgenic soybean plot. Gene flow was observed up to 10.0 m from the transgenic soybean plot at a rate of 0.049%.
In total, 54,554 progenies from non-transgenic soybean planted with CT-2062 were assessed to identify hybrids (Table 4). The presence of the 114-bp igf-1 gene fragment in 27 samples was confirmed by PCR analysis, and the overall gene flow rate was 0.052%. The highest rate (0.289%) was also observed for the soybean planted at the closest distance (0.5 m) from the transgenic soybean plot. Gene flow was observed up to 5.0 m from the transgenic soybean plot.
Non-transgenic soybean planted with CT-4025 had 52,337 progenies (Table 4). Among them, the 185-bp trx gene fragment was observed in 23 samples, and the overall gene flow rate was 0.044%. The highest rate (0.464%) was observed at the closest distance from the transgenic soybean plot. Here, 13 of the 2801 progenies were hybrids between transgenic and non-transgenic soybeans. Gene flow was observed up to 13.1 m from the transgenic soybean plot.
From the seeds collected from the 19th row (9.5 m distance from the transgenic soybean plot) of non-transgenic soybean north-east plots planted with CT-2062, we found two glufosinate-resistant progenies. When we analyzed the samples using primers that could detect the igf-1 gene (for CT-2062), the igf-1 gene was not detected among these samples. Therefore, we tested other primers that could detect either the egf (for CT-1001) or trx (for CT-4025) gene and found that the trx gene was present in these samples. Because we pooled the seeds collected from the five non-transgenic soybeans planted on each row with the same distance from the transgenic soybean plot, it was not possible to locate the exact position of collection. Therefore, we estimated the distance by drawing a hypothetical line between the central edge of the transgenic soybean plot and the center of the 19th row where the hybrid progenies were found (Fig. 1).
Discussion
Pollen characteristics, including pollen grain size and viability, are related to the potential for hybridization and are a major component in risk assessment of transgenic crops [50, 51]. The lack of significant differences in pollen grain size, viability determined by staining, and pollen germination rates between transgenic soybean events and the non-transgenic parental cultivar in our study indicates that the overexpression of EGF, IGF-1, or TRX did not alter pollen characteristics in soybean. The mean pollen grain diameter of non-transgenic and transgenic soybean in our study ranged from 24.8 to 27.3 μm. Kaltchuk-Santos et al. [52] reported that the pollen grain diameter of four soybean cultivars ranged from 25.23 to 26.85 μm, which was similar to that in our study. Horak et al. [53] reported that pollen grain diameter of transgenic and non-transgenic soybean ranged from 21.6 to 23.7 μm, which was slightly smaller than that found in our study. Mean pollen diameters of soybean as large as 27.3 μm and 30.4 μm are also reported [10].
Flowering phenologies of transgenic soybean events were not different from those of non-transgenic soybean, and flowering periods were overlapped in our study. We found that the highest gene flow rates were 0.21%, 0.29%, and 0.46% for transgenic events CT-1001, CT-2062, and CT-4025, respectively, at the closest distance (0.5 m) from transgenic soybean plots. This result is similar to that of the studies that found 0.26 to 0.45% of the highest gene flow rate at 0.5 m distance from the transgenic soybean plots [20, 28]. At a distance of 0.7 to 1 m from the transgenic soybean pollen source, the highest gene flow rates of 0.03 to 0.52% were also observed [21, 26, 29, 31]. The highest gene flow rates reported in studies using transgenic soybean were mostly below 0.6%.
The gene flow rates were considerably reduced in the second rows, at a 1.0 m distance from the transgenic soybean plots in our study. The maximum distance that gene flow could occur from the pollen source in our study was 5 m, 10 m, and 13.1 m for CT-2062, CT-1001, and CT-4025, respectively.
The farthest distance that transgene flow can occur was reported as 0.7 to 10 m [21, 22, 25, 26, 29,30,31]. However, distance as far as 15 m [28] and 29 m [27] have been reported in China. Caviness [4] also reported an outcrossing of 0.01% between soybean cultivars at 10 to 15.5 m distance. Our data fell within the ranges reported in the literature and indicate that an isolation distance greater than at least 13 m from transgenic soybean is required to prevent within-crop gene flow in soybean.
Inefficient transgene biocontainment can incur great economic losses, as seen in the StarLink incident; moreover, pollen-mediated gene flow is a primary mode of unwanted biocontamination [54]. Determining the isolation distance between transgenic crops and sexually compatible crops is one of the various measures, including using pollen traps and buffer zones, to prevent pollen flow into neighboring fields [55]. Conducting confined field trials of transgenic crops is essential to obtain such data for decision making.
In summary, we observed that rates of gene flow from three transgenic soybean events, developed to obtain recombinant proteins (EGF, IGF-1, or TRX) for use in the skin care industry, to the non-transgenic parental cultivar in the field and the farthest distance at which gene flow can occur fell within the reported ranges. Pollen characteristics were not significantly different between non-transgenic and transgenic soybean events. As the potential markets for transgenic crops as a recombinant protein factory increase, gene flow from transgenic to non-transgenic conventional crops will become a key decision factor for policy makers during the approval process of transgenic crops. Our study may provide useful baseline data for the prevention of transgenic soybean seed contamination caused by transgene flow.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Woodworth CM (1922) The extent of natural cross-pollination in soybeans. J Am Soc Agron 14:278–283
Garber RJ, Odland TE (1926) Natural crossing in soybeans. J Am Soc Agron 18:967–970
Takagi M (1926) On the frequency of the spontaneous hybridization in soybean. Annals Agric Exp Stn Gov Gen Chosen 4:323–324
Caviness CE (1966) Estimates of natural cross-pollination in Jackson soybeans in Arkansas. Crop Sci 6:211–212
Chiang YC, Kiang YT (1987) Geometric position of genotypes, honeybee foraging patterns and outcrossing in soybean. Bot Bull Acad Sinica 28:1–11
Gumisiriza G, Rubaihayo PR (1978) Factors that influence outcrossing in soybean. J Agron Crop Sci 147:129–133
Ray JD, Kilen TC, Abel CA, Paris RL (2003) Soybean natural cross-pollination rates under field conditions. Environ Biosaf Res 2:133–138
Beard BH, Knowles PF (1971) Frequency of cross-pollination of soybeans after seed irradiation. Crop Sci 11:489–492
Brim CA, Young MF (1971) Inheritance of a male-sterile character in soybeans. Crop Sci 11:564–566
Yoshimura Y (2011) Wind tunnel and field assessment of pollen dispersal in soybean [Glycine max (L.) Merr.]. J Plant Res 124:109–114
Ortiz-Perez E, Mian RMA, Cooper RL, Mendiola T, Tew J, Horner HT, Hanlin SJ, Palmer RG (2008) Seed-set evaluation of four male-sterile, female-fertile soybean lines using alfalfa leafcutting bees and honey bees as pollinators. J Agr Sci 146:461–469
Milfont MO, Rocha EEM, Lima AON, Freitas BM (2013) Higher soybean production using honeybee and wild pollinators, a sustainable alternative to pesticides and autopollination. Environ Chem Lett 11:335–341
Gill KA, O’Neal ME (2015) Survey of soybean insect pollinators: community identification and sampling method analysis. Environ Entomol 44:488–498
Dai J, Zhang R, Wei B, Nie Z, Xing G, Zhao T, Yang S, Gai J (2017) Key biological factors related to outcrossing-productivity of cytoplasmic-nuclear male-sterile lines in soybean [Glycin max (L.) Merr.]. Euphytica 213:266
Bletter DC, Fagúndez GA, Caviglia OP (2018) Contribution of honeybees to soybean yield. Apidologie 49:101–111
Weber CR, Hanson WD (1961) Natural hybridization with and without ionizing radiation in soybeans. Crop Sci 1:389–392
Boerma HR, Moradshahi A (1975) Pollen movement within and between rows to male-sterile soybeans. Crop Sci 15:858–861
Ahrent DK, Caviness CE (1994) Natural cross-pollination of twelve soybean cultivars in Arkansas. Crop Sci 34:376–378
ISAAA (2018) Global status of commercialized biotech/GM crops in 2018: Biotech crops continue to help meet the challenges of increased poulation and climate change. ISAAA Brief No. 54. ISAAA, Ithaca
Abud S, de Souza PIM, Moreira CT, Andrade SRM, Ulbrich AV, Vianna GR, Rech EL, Aragão FJL (2003) Gene flow in transgenic soybean in the Cerrado region, Brazil. Pesq Agropec Bras 38:1229–1235
Abud S, de Souza PIM, Vianna GR, Leonardecz E, Moreira CT, Faleiro FG, Júnior JN, Monteiro PMFO, Rech EL, Aragão FJL (2007) Gene flow from transgenic to nontransgenic soybean plants in the Cerrado region of Brazil. Genet Mol Res 6:445–452
Pereira WA, Sávio FL, dos Dias DCF, Cruz CD, Borém A (2012) Reciprocal gene flow between conventional and genetically modified soybean cultivars. Pesq Agropec Bras 47:227–236
Lemes ES, Barros ACSA, Peske ST, de Tunes LM, Levien OLM (2014) Estimate of the gene flow between transgenic and nontransgenic soybean cultivars. Appl Res Agrotechnol 7:97–102
Liu Q, Li X, Liu Z, Li T, Lei B (2008) Study on Roundup Ready soybean’s Roundup Ready gene flowing I. Study on Roundup Ready gene move to soybean by anemophily. Heilongj Agric Sci 1:14–16
Lü X, Wang H, Liu Q, Zhao G, Li X, Xu G, Zhang L, Liu J, Liu Z, Li N, Li T, Lei B (2009) Biosafety of Roundup Ready soybean (RRS) planted in black soil ecosystem. Soybean Sci 28:260–266
Zhang B, Li Y, Gai J, Li W (2011) Distance and frequency of gene flow in transgenic soybean overexpressing TaDREB3. Soybean Sci 30:563–565
Liu J, Zhou B, Yang C, Li Y, Jiang L, Zhang M, Tao B, Qiu L (2012) Gene flowing of genetically modified glyphosate-resistant soybean with EPSPS. Soybean Sci 31:517–521
Huang WK, Peng H, Wang GF, Cui JK, Zhu LF, Long HB, Peng DL (2014) Assessment of gene flow from glyphosate-resistant transgenic soybean to conventional soybean in China. Acta Physiol Plant 36:1637–1647
Yoshimura Y, Matsuo K, Yasuda K (2006) Gene flow from GM glyphosate-tolerant to conventional soybeans under field conditions in Japan. Environ Biosaf Res 5:169–173
Lee B, Oh SD, Chang A (2018) Influence of gene flow from GM to non-GM soybeans by the size of the pollen donor. Kor J Agric Sci 45:591–600
Kim HJ, Kim DY, Moon YS, Pack IS, Park KW, Chung YS, Kim YJ, Nam KH, Kim CG (2019) Gene flow from herbicide resistant transgenic soybean to conventional soybean and wild soybean. Appl Biol Chem 62:54
Ma JKC, Drake PMW, Christou P (2003) The production of recombinant pharmaceutical proteins in plants. Nat Rev Genet 4:794–805
Lau OS, Sun SSM (2009) Plant seeds as bioreactors for recombinant protein production. Biotechnol Adv 27:1015–1022
Magnusdottir A, Vidarsson H, Björnsson JM, Örvar BL (2013) Barley grains for the production of endotoxin-free growth factors. Trends Biotechnol 31:572–580
Xu J, Dolan MC, Medrano G, Cramer CL, Weathers PJ (2012) Green factory: plants as bioproduction platforms for recombinant proteins. Biotechnol Adv 30:1171–1184
He Y, Schmidt MA, Erwin C, Guo J, Sun R, Pendarvis K, Warner BW, Herman EM (2016) Transgenic soybean production of bioactive human epidermal growth factor (EGF). PLoS ONE 11:e0157034
Anderson EJ, Ali ML, Beavis WD, Chen P, Clemente TE, Diers BW, Graef GL, Grassini P, Hyten DL, McHale LK, Nelson RL, Parrott WA, Patil GB, Stupar RM, Tilmon KJ (2019) Soybean [Glycine max (L.) Merr.] breeding: History, improvement, production and future opportunities. In: Al-Khayri JM, Jain M, Johnson DV (eds) Advances in plant breeding strategies: legumes. Springer, Cham, pp 431–516
Cohen S, Carpenter G (1975) Human epidermal growth factor: isolation and chemical and biological properties. Proc Nat Acad Sci USA 72:1317–1321
Carpenter G, Cohen S (1990) Epidermal growth factor. J Biol Chem 265:7709–7712
Rinderknecht E, Humbel RE (1978) The amino acid sequence of human insulin-like growth factor I and its structural homology with proinsulin. J Biol Chem 253:2769–2776
Laron Z (2001) Insulin-like growth factor 1 (IGF-1): a growth hormone. J Clin Pathol Mol Pathol 54:311–316
Holmgren A (1989) Thioredoxin and glutaredoxin systems. J Biol Chem 25:13963–13966
Zhou JD, Wang CX, Wu JL, Fukunaga A, Cheng ZS, Wang JQ, Yamauchi A, Yodoi J, Tian H (2020) Anti-allergic and anti-inflammatory effects and molecular mechanisms of thioredoxin on respiratory system diseases. Antioxid Redox Sign 32:785–801
Amin MR, Oh SD, Suh SJ (2020) Comparing the effects of GM and non-GM soybean varieties on non-target arthropods. Entomol Res 50:423–432
Kim DY, Eom MS, Kim HJ, Pack IS, Park JH, Park KW, Nam KH, Oh SD, Kim JK, Seo JS, Kim CG (2020) Assessing invasiveness of genetically modified soybean expressing human epidermal growth factor gene. Weed Turf Sci 9:119–128
Ha TJ, Lim SG, Shin SH, Choi KJ, Baek IY, Lee SC, Park KY, Shin SO (2009) Maturity grouping of Korean soybean cultivars and character relationships according to the planting date. Kor J Crop Sci 54:104–118
Salem MA, Kakani VG, Koti S, Reddy KR (2007) Pollen-based screening of soybean genotypes for high temperatures. Crop Sci 47:219–231
Korea Meteorological Administration (2017) Annual climatological report. Korea Meteorological Administration, Seoul
Zaiontz C (2019) Real Statistics Using Excel. http://www.real-statistics.com
Shibaike H, Matsuo K (2007) Pollen dispersal and hybridization of genetically modified crops. In: Asai M, Shibaike H (eds) Weed ecology and agroecosystems—introduced plants and genetically modified crops. Bun-ichi Sogo Shuppan Co., Tokyo, pp 219–245
EFSA (European Food Safety Authority) Panel on Genetically Modified Organisms (2010) Guidance on the environmental risk assessment of genetically modified plants. EFSA J 8:1879
Kaltchuk-Santos E, Zanettini MHB, Mundstock E (1993) Pollen dimorphism in soybean. Protoplasma 174:74–78
Horak MJ, Rosenbaum EW, Kendrick DL, Sammons B, Philips SL, Nickson TE, Dobert RC, Perez T (2015) Plant characterization of Roundup Ready 2 Yield® soybean MON 89788, for use in ecological risk assessment. Transgenic Res 24:213–225
Clark M, Maselko M (2020) Transgene biocontainment strategies for molecular farming. Front Plant Sci 11:210
Menrad K, Hirzinger T, Reitmeier D (2011) Coexistence and traceability of GMOs in the agro-food sector. In: Baram M, Bourrier M (eds) Governing risk in GM agriculture. Cambridge University Press, New York, pp 139–168
Acknowledgements
Not applicable.
Funding
This research was supported by grants from the KRIBB Research Initiative Program and “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01368601)”, Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
DYK, MSE, KWP, and CGK analyzed the data and wrote the manuscript; DYK, MSE, HJK, EMK, ISP, KHN performed the experiments; JSS prepared the plant materials; JHP, SDO, JKK, JSS, and CGK designed the research. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, D.Y., Eom, M.S., Kim, H.J. et al. Gene flow from transgenic soybean, developed to obtain recombinant proteins for use in the skin care industry, to non-transgenic soybean. Appl Biol Chem 63, 65 (2020). https://doi.org/10.1186/s13765-020-00550-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-020-00550-w