Abstract
Berry fruits have attracted attention because of their purported benefits for aging, cardiovascular disease, and cancer. Therefore, highly priced berry fruits might be targets for food adulteration and fraud. In this study, eight species-specific primer sets based on the single nucleotide polymorphism of the chloroplast genomes of four berry fruits (aronia, blackberry, cranberry, and strawberry) were developed for quantitative real-time PCR (qPCR) analysis by SYBR Green staining with the aim of preventing berry fruit food fraud. The developed primer pairs exhibited high efficiencies ranging from 88 to 110% with strong correlation coefficients (R2 > 0.99) for the amplification of each target species. However, no clear correlation coefficients were evident for non-target species. To evaluate the practicality of the developed primers, 18 commercial berry fruit products were verified by qPCR analysis. The developed primer pairs were amplified to a low Ct value (range 16.1–23.3) for the target species and proved capable of detecting target species in berry fruit commercial foods. Therefore, the developed qPCR-based species-specific markers could be suitable for the prevention of berry fruit food fraud and to verify marker reliability.
Similar content being viewed by others
Introduction
Berry fruits have been implicated in the lessening or prevention of diseases, and account for a large percentage of the fruits consumed as part of the human diet [1]. Berry fruits are also widely used in processed and derived products, including dried and canned fruits, yogurt, drinks, jams and jellies, as well as fresh and frozen fruits [2]. It has been amply demonstrated that berry fruits contain antioxidant and anti-inflammatory polyphenols, such as anthocyanins and phenolic acids, which play an important role in aging [3] and the prevention and treatment of cardiovascular disease [4] and cancer [5]. Therefore, consumption of berry fruits has been rapidly increasing worldwide.
Food fraud, which exploits food buyers for economic benefit, has a long history. The United States Grocery Manufacturers Association estimates that food fraud costs the global food industry $10–$15 billion annually, affecting about 10% of all sold commercial foods [6]. In response, various technologies incorporating molecular markers and biochemical analysis as two examples have been developed for prevent food fraud involving plants, such as saffron [7], orange [8], Orostachys japonica [9], and Cynanchum wilfordii [10]. However, prevention of food fraud for berry fruits is relatively unexplored. In general, chemical analysis is time-consuming and has limitations related to repeatability and reproducibility [11]. As an alternative to chemical assays, DNA-based assays are considered excellent tools for the identification of plant species in commercial foods. However, research on the classification of berry fruit based on DNA is also scant. DNA-based methods for detecting contamination with berry fruits in commercial foods are needed.
Chloroplast genes are generally present in hundreds of copies per cell, and chloroplast genomes are protected from decomposition during food processing since they are enclosed by two membrane layers [12]. Thus, chloroplast genomes have been widely used in plant species identification to develop species-specific detection methods [13] as well as in evolutionary studies. In particular, the chloroplasts maturase K (matK) and RubisCO large-subunit genes (rbcL) have proven to be excellent for DNA barcoding-based species identification [14, 15]. The use of DNA barcoding for plant species identification has been studied [16, 17]. In addition, the trnL–trnF region is frequently used along with matK and rbcL for DNA barcoding-based species identifications due to the extensive polymorphisms among species [18, 19].
DNA-based polymerase chain reaction (PCR) analyses have been widely used for preventing food fraud because of its economic and timesaving advantages compared to biochemical analyses. In particular, the high accuracy and sensitivity of quantitative real-time PCR (qPCR) assays could enable the detection of very low levels of target DNA in commercial foods. In this study, we developed four berry fruit species-specific molecular markers using DNA polymorphisms of chloroplast genes with the aim of utilizing the markers to prevent food fraud, and verified their application in commercial berry fruit food products.
Materials and methods
Sample preparation
Four species of berry fruits were used (Table 1): aronia (Aronia melanocarpa), blackberry (Rubus fruticosus), cranberry (Vaccinium macrocarpon), and strawberry (Fragaria × ananassa). All were purchased from Korea plant nursery (http://www.treeinfo.com, Taean, Korea). A total of 18 commercial berry fruit products were purchased from local or oversea markets (Table 1).
Genomic DNA extraction
Genomic DNAs of all fruit leaves and commercial products were extracted using the i-genomic Plant DNA Extraction Mini Kit (iNtRON Biotechnology, Seongnam, Korea) according to the manufacturer`s protocol. Extracted genomic DNA concentration was measured using a Qubit® 2.0 Fluorometer (Invitrogen, Life Technologies, Grand Island, NY, USA) with a Qubit dsDNA BR Assay Kit (Invitrogen). The amplification capacity of the extracted DNA was assessed with universal plant primer pairs that target a conserved 18S rRNA nuclear region [20].
Sequence alignment and primer design
Three chloroplast gene regions (matK, rbcL, and trnL-F) were used to develop species-specific molecular markers. All berry fruit chloroplast sequences were downloaded from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). Downloaded chloroplast sequences were aligned using ClustalW2 (ftp://ebi.ac.uk/pub/software/clustalw2/) and Software Bio-edit 7.2 (Ibis Biosciences, Carlsbad, CA, USA). The species-specific primer pairs were designed using Beacon Designer™ (PRIMER Biosoft, Palo Alto, CA, USA). All designed primer sets were synthesized by a commercial service (Macrogen, Seoul, Korea). The primer sequences used in this study are listed in Table 2.
Real-time PCR analysis
qPCR was carried out in a final volume of 20 µL using a CFX Connect™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) with SYBR Green dye. The reaction mixture consisted 10 µL of SYBR® Green TOP real qPCR 2xPreMIX (Enzynomics™, Daejeon, Korea), 10 ng of genomic DNA and 10 pmol of each primer sets, with adjustment to a final volume of 20 μL with PCR-grade water. The experiment conditions were as follows: 10 min at 95 °C, followed by 45 cycles of 10 s at 95 °C, annealing at the appropriate annealing temperature (Tm) and time of each primer pair, and 30 s at 72 °C. The PCR products were denatured at 95 °C for 10 s and then annealed at 65 °C for 5 s. This step was followed by a melt-curve analysis at temperatures ranging from 60 to 95 °C. For sensitivity analysis, fruit leaf DNAs of each species were diluted tenfold into five series (0.001–10 ng/μL) and used for real-time PCR. In addition, extracted each commercial product DNAs was quantitated to 10 ng and used for real-time PCR.
Cloning of PCR amplicons
To identity PCR products amplified from the correct target regions, they were cloned using commercial product RBC T&A Cloning Vector (Real Biotech Corporation, Taipei, Taiwan) and a ligation mix (TaKaRa Bio, Shiga, Japan) according to the manufacturer’s protocol. Conventional PCR was performed using TaKaRa Ex Taq™ DNA polymerase (TaKaRa Bio). PCR reaction was performed with an initial denaturation 5 min at 95 °C, followed by 35 cycles of 10 s at 95 °C, 30 s at Tm °C (each primer), 1 min at 72 °C, and finally 5 min at 72 °C. Then, the amplicon was cloned into the T&A Vector and plasmid DNA was purified using the Plasmid Mini-Prep Kit (Elpis Biotech, Daejeon, Korea). Nucleotide sequences were analyzed by a commercial service (Macrogen).
Determination of amplification efficiency, correlation coefficient, and limit of detection (LOD)
To evaluate the correlation between DNA concentration and cycle threshold (Ct), standard curves were obtained using tenfold diluted DNA samples of the four berry fruits species at concentrations of 0.001–10 ng. The correlation coefficient (R2) was determined by using the linear regression method (R2 ≥ 0.98) [21]. The amplification efficiency was calculated on the basis of the standard curve using the equations E = 10−1/slope, and efficiency (%) = (E−1) × 100. The LOD was regarded as the analytical concentration at which the method detected the presence of a target gene in at least 95% of true-positive biological samples (< 5% of false-negative results) [22].
Results and discussion
Species-specific qPCR primers design
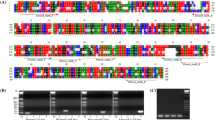
Three chloroplast gene sequences (matK, trnL-F, and rbcL) of the four berry fruits were obtained from NCBI. The chloroplast sequences were aligned using ClustalW2 to identify single nucleotide polymorphisms (SNPs) or InDels (insertions and deletions) among the four berry fruits. A host of SNPs and/or InDels were found in each of three genes among the sequences (Additional file 1: Fig. S1), suggesting that the differences could be useful for designing the species-specific primers.
Species-specific primers were designed using a commercial program based on SNPs or InDels of the berry fruit cpDNA sequences. Since DNA of foods tends to be degraded into short fragments during food processing steps, such as heat treatment, small PCR products are better amplified compared to large PCR products in processed foods [23]. Therefore, we designed eight species-specific primer pairs that could amplify short amplicons of 94–224 bp to detect target species in processed foods (Table 2). All species-specific primers were designed based on two or more SNPs to clearly distinguish each berry fruit (Additional file 1: Fig. S1).
Evaluation of the amplification efficiency and sensitivity of the species-specific primers
We evaluated the efficiency and sensitivity of the developed primers for qPCR analyses using tenfold serially diluted DNA (10–0.001 ng) of each fruit and examining individual statistical measurements using a regression analysis. The quality of extracted DNAs of each fruits was first evaluated by qPCR using universal plant primer pairs [20]. The 18S rRNA was efficiently amplified in all samples, with Ct values in the range of 14.12–14.35. Then, efficiencies of all species-specific primers were confirmed to be in the range of 88–108% with a strong correlation coefficient (R2 > 0.99) for the target species (Table 3). In addition, the slopes of the linear equations for the target species ranged from − 3.13 to − 3.63. However, the correlation coefficient of non-target (other) species was not evident in all the developed primer pairs (Fig. 1). These results suggest that the developed primer pairs could be very suitable for detecting the target species. To confirm whether the PCR amplicons were derived from the correct target regions, the products were cloned and sequenced. No differences were found between both sequences NCBI deposited and amplified by the primer (Additional file 1: Fig. S2). Subsequently, target-specificities of the developed primers were evaluated using end-point PCR (30 cycles) and the amplicons were visualized by agarose gel electrophoresis (Fig. 2a). All PCR products were amplified with each of the desired size in the genomic DNA. In addition, sharp peaks were obvious for the target species in PCR products amplified with each of the developed primer pairs (Fig. 2b). These findings suggest that the develop primer pairs could be useful for distinguishing between the four berry fruit species.
Standard curves obtained by analyzing serially diluted DNAs of the four berry fruits. The standard curves were obtained on the basis of efficiency and correlation of coefficient (R2) of DNA extracted from the plant leaves. The x-axis and the y-axis represent log DNA concentration and Ct value, respectively
Application of the primers to commercial food products of berry fruits
To gauge the applicability of the developed primer pairs, 18 commercial products were purchased from local or oversea markets and then tested. Prior to applying the commercial foods, the universal primers were used to test whether the DNA extracted from the food products were suitable for the qRT-PCR assay. Results of the qPCR analysis with the universal plant primer pairs (18S rRNA amplification) showed low Ct values in the range of 12.83–17.15 for all products, indicating that the quality of the extracted DNA was suitable for further assays.
Based on qPCR results, Ct values of the 18 commercial products were determined (Table 4). All Ct values amplified with target primers were lower than the LOD (10 pg Ct values). However, Ct values with non-target primers were higher than the LOD. Our results for all 18 commercial products were consistent with the indicated ingredients, suggesting that the developed primer pairs would be useful to detect the target berry species in commercial foods.
Fruits are commonly processed to fruit juices. The market sectors for fruit juices have been rapidly growing. Therefore, highly priced fruit juices have been targets for food adulteration and fraud [24]. Since the most frequent profit-procedures are simple dilution with water, the addition of sugar or cheap alternatives, a host of non-targeted high-performance liquid chromatography-mass spectrometry metabolomics fingerprinting techniques coupled with chemometric analysis have been developed for the juice-type food authenticity testing [24]. Since the impact of berry fruits intake on human health, performance, and disease as superfoods have been firmly established, they have been commonly consumed worldwide in fresh and processed forms, such as dried powders and teas [5]. Therefore, alternative detection technologies would be useful for detection of berry fruit adulteration and fraud.
SYBR Green-based qPCR analysis is a useful tool for the detection and quantification of species-specific nucleotides. This method is faster and more stable than other chemical assays [25]. SYBR Green-based techniques can detect other plant species in processed foods, such as hazelnuts [26], almonds [27], DNA allergens [28], and C. wilfordii and C. auriculatum [10]. In this study, we report the development of a DNA-based SYBR Green qPCR assay for rapid and sensitive species-specific detection (up to 10 pg DNA) of four berry fruits. In additional, the markers were applied successfully to 18 commercial berry fruit foods. The developed molecular markers could be useful for detection of food fraud and adulteration in commercial berry fruits foods markets.
References
Bazzano LA (2005) Dietary intake of fruit and vegetables and risk of diabetes mellitus and cardiovascular diseases. WHO, Geneva
Seeram NP (2006) Berries. Nutritional oncology, 2nd edn. Elsevier, Amsterdam, pp 615–628
Thangthaeng N, Poulose SM, Miller MG, Shukitt-hale B (2016) Preserving brain function in aging: the anti-glycative potential of berry fruit. NeuroMol Med 18(3):465–473
Vendrame S, Del Bo C, Ciappellano S, Riso P, Klimis-zacas D (2016) Berry fruit consumption and metabolic syndrome. Antioxidants 5(4):34
Seeram NP (2008) Berry fruits for cancer prevention: current status and future prospects. J Agric Food Chem 56(3):630–635
Johnson R (2014) Food Fraud and "Economically Motivated Adulteration" of Food and Food Ingredients, Congressional Research Service, Washington, D.C.
Villa C, Costa J, Oliveira MBP, Mafra I (2017) Novel quantitative real-time PCR approach to determine safflower (Carthamus tinctorius) adulteration in saffron (Crocus sativus). Food Chem 229:680–687
Han J, Wu Y, Huang W, Wang B, Sun C, Ge Y, Chen Y (2012) PCR and DHPLC methods used to detect juice ingredient from 7 fruits. Food Control 25(2):696–703
An J, Moon J, Jang CS (2018) Markers for distinguishing Orostachys species by SYBR Green-based real-time PCR and verification of their application in commercial O. Japonica food products. Appl Biol Chem 61(5):499–508
Kim JH, Moon J, Kang TS, Kwon K, Jang CS (2017) Development of cpDNA markers for discrimination between Cynanchum wilfordii and Cynanchum auriculatum and their application in commercial C. wilfordii food products. Appl Biol Chem 60(1):79–86
Yalcinkaya B, Yumbul E, Mozioglu E, Akgoz M (2017) Comparison of DNA extraction methods for meat analysis. Food Chem 221:1253–1257
Garino C, De Paolis A, Coisson JD, Bianchi DM, Decastelli L, Arlorio M (2016) Sensitive and specific detection of pine nut (Pinus spp.) by real-time PCR in complex food products. Food Chem 194:980–985
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16(3):142–147
CBOL Plant WORKING GROUP (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106(31):12794–12797
Li F, Kuo L, Rothfels CJ, Ebihara A, Chiou W, Windham MD, Pryer KM (2011) rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS ONE 6(10):e26597
Asahina H, Shinozaki J, Masuda K, Morimitsu Y, Satake M (2010) Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J Nat Med 64(2):133–138
Burgess KS, Fazekas AJ, Kesanakurti PR, Graham SW, Husband BC, Newmaster SG, Percy DM, Hajibabaei M, Barrett SC (2011) Discriminating plant species in a local temperate flora using the rbcL matK DNA barcode. Methods Ecol Evol 2(4):333–340
Cabelin VL, Alejandro GJ (2016) Efficiency of matK, rbcL, trnH-psbA, and trnL-F (cpDNA) to molecularly authenticate philippine ethnomedicinal apocynaceae through DNA barcoding. Pharmacogn Mag 12(Suppl 3):S384–S388
De Groot GA, During HJ, Maas JW, Schneider H, Vogel JC, Erkens RH (2011) Use of rbcL and trnL-F as a two-locus DNA barcode for identification of NW-European ferns: an ecological perspective. PLoS ONE 6(1):e16371
Allmann M, Candrian U, Hffelein C, Lthy J (1993) Polymerase chain reaction (PCR): a possible alternative to immunochemical methods assuring safety and quality of food. Detection of wheat contamination in non-wheat food products. Z Lebensm Unters Forsch 196:248–251
Ramakers C, Ruijter JM, Deprez RHL, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339(1):62–66
Ferreira T, Farah A, Oliveira TC, Lima IS, Vitório F, Oliveira EM (2016) Using real-time PCR as a tool for monitoring the authenticity of commercial coffees. Food Chem 199:433–438
Lo Y, Shaw P (2018) DNA-based techniques for authentication of processed food and food supplements. Food Chem 240:767–774
Hong E, Lee SY, Jeong JY, Park JM, Kim BH, Kwon K, Chun HS (2017) Modern analytical methods for the detection of food fraud and adulteration by food category. J Sci Food Agric 97(12):3877–3896
Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, Bell SM, Combaret V, Puisieux A, Mighell AJ (2003) Real-time PCR based on SYBR-green I fluorescence: an alternative to the TaqMan assay for a relative quantification of gene rearrangements, gene amplifications and micro gene deletions. BMC Biotechnol 3(1):18
Arlorio M, Cereti E, Coisson J, Travaglia F, Martelli A (2007) Detection of hazelnut (Corylus spp.) in processed foods using real-time PCR. Food Control 18(2):140–148
Pafundo S, Gulli M, Marmiroli N (2009) SYBR® GreenER™ real-time PCR to detect almond in traces in processed food. Food Chem 116(3):811–815
Pafundo S, Gullì M, Marmiroli N (2010) Multiplex real-time PCR using SYBR® GreenER™ for the detection of DNA allergens in food. Anal Bioanal Chem 396(5):1831–1839
Authors’ contributions
CSJ conceived of the overall study. JA, J-CM, and CSJ co-wrote the paper. JA, GSK, and JHK performed the experiments and analyzed the data. All authors read and approved the final manuscript.
Acknowledgements
This research was supported by a Grant (17162MFDS065) from Ministry of Food and Drug Safety in 2018.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Figure S1.
Alignments of three chloroplast gene sequences of Aronia melanocarpa (Aronia), Rubus fruticosus (Blackberry), Vaccinium macrocarpon (Cranberry), and Fragaria × ananassa (Strawberry) using ClustalW2 program. Designed species-specific primers are represented by arrows and include Red for aronia; black for blackberry; green for cranberry; and blue for strawberry. (A) matK gene, (B) rbcL gene, and (C) trnL-F gene. Figure S2. Comparison of nucleotide sequences between P the NCBI database and PCR amplicons produced by species-specific primers. (A) Aronica matK primer; (B) Blackberry trnL-F primer; (C) Cranberry matK primer; and (D) Strawberry trnL-F primer.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
An, J., Moon, JC., Kim, J.H. et al. Development of DNA-based species-specific real-time PCR markers for four berry fruits and their application in commercial berry fruit foods. Appl Biol Chem 62, 10 (2019). https://doi.org/10.1186/s13765-019-0413-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13765-019-0413-9