Abstract
Colibacillosis is a major disease affecting poultry leads to high morbidity and mortality which causing tremendous economic losses worldwide. These economic disparities are amplified among low and middle-income where sanitation and hygiene are challenged by the increasing demand for quality sources of animal protein. With a view to investigating the prevalence of virulence genes and QACs resistance genes as well as monitoring the antibiogram of E. coli strains, a total of 368 specimens were collected from diseased broiler chickens (n = 226) and environmental sources (n = 142) at large-scale poultry farms in Ismailia Governorate, Egypt. The bacteriological examination proved that E. coli prevalence was 26.76% and 50.44% in the farm environment and diseased broilers, respectively. In tandem, the isolated E. coli strains were serogrouped, determining the most common serotypes were O78, O1:H7, O91:H21 and O126. Isolates were tested for antimicrobial susceptibility against 12 antibiotics, screened for 4 virulence genes (iss, papC, eaeA, and cfaI), and screened for 3 QACs resistance genes (qacEΔ1, qacA/B, and qacC/D). All the tested strains were positive for iss and papC genes, only 20.3% of the tested strains were positive for eaeA gene, moreover, the examined strains were negative to CFAI gene. Furthermore, all the tested strains were positive for qacEΔ1, qacA/B, and qacC/D genes. In conclusion; virulence genes (iss, papC) as well as QACs resistance genes are common in avian Pathogenic E. coli and environmental strains and are mainly associated with multi-drug resistance phenomena.

Similar content being viewed by others
Introduction
The rise of poultry production and industrial breeds of chicken, such as meat broilers, have been instituted as a method to promote gender equity, economic stability, and food security within many low and middle-income countries (LMICs). Despite the benefits, large-scale poultry production facilities within LMICs are often confronted with a tradeoff between animal welfare and addressing a growing economic demand, leading to high amounts of sub-therapeutic antibiotics for growth promotion and prophylaxis as well as disinfectant agents (Joint 2008; Udomsantisuk et al. 2011). High use of antibiotics and disinfectants could promote further antibiotic resistance (ARB) and disinfectant resistance (DR) (Eid et al. 2016).
One primary target for antibiotic and disinfect use is colibacillosis, which remains one of the major drivers of poultry morbidity and mortality, leading to severe losses (Barnes et al. 2008). These biosecurity risks can be further intensified as some E. coli avian diseases can be zoonotically transmitted via a trophic transmission (González-Zorn et al. 2005) or occupational exposure (Bisi-Johnson et al. 2011). Knowledge of E. coli serology can inform how to best treat diseases based upon typing categorizations. For instance, E. coli serotypes can cause intestinal illness with digestive signs, while other serotypes referred to as Avian Pathogenic E. coli belong to ExPEC that cause various symptoms in chicken either systemic or localized including; omphalitis, respiratory colibacillosis and colisepticemia (Rodriguez-Siek et al. 2005; Mellata 2013). The plasticity of E. coli pathogenicity is a result of an extensive range of virulence factors that are regulated and encoded by virulence determinant genes such as (iss, papC, eaeA, and CFAI) (De Carli et al. 2015; Eid et al. 2019). Studies have found links of co-selection for bacterial resistance from disinfectant and antibiotic use. Often, selection for resistance to antibiotics can inadvertently lead to drug resistance by movable genetic components (Noguchi et al. 2005; Chuanchuen et al. 2007). Additionally, these multimodal pathways for resistance can also promote increased pathogenicity in other species of bacteria through horizontal gene transfer. The quaternary ammonium compounds could be a major cause of the antibacterial cross-resistance development (Buffet-Bataillon et al. 2012). Various disinfectant resistant genes were recorded in multidrug-resistant pathogenic bacterial species (Zhang et al. 2015) including; qacA/B, qacC/D, qacE, and qacG genes (Correa et al. 2008). Often, in low-to- middle large-scale animal operations often apply high amounts of disinfectants and sub-therapeutic antibiotics to mitigate are chemical agents that used to kill microorganisms on inanimate instruments by various mechanisms as well as a wide spectrum of activity and potency (Fraise et al. 2013). Quaternary ammonium compounds are less toxic, non-irritating substances that are widely used to disinfect poultry farms environment (Bore et al. 2007). Many reports revealed a molecular relationship between qac genes and antibiotic resistance in certain pathogenic bacteria (Sidhu et al. 2001).
This study was aimed to investigate the prevalence of virulence genes (iss, papC, eaeA, and CFAI) and QACs resistance genes (qacEΔ1, QacA/B, and QacC/D) in E. coli strains originated from diseased broiler chicken and farm environment as well as monitoring of the antimicrobial susceptibility of the isolated strains.
Materials and methods
Sampling
A total of 368 specimens were collected aseptically from large scale farms [142 environmental samples: feeders (n = 32), drinkers (n = 32), walls (n = 36) and floors (n = 42) and 226 samples from diseased broiler chickens: heart (n = 70), liver (n = 82), lung (n = 12), yolk (n = 30), spleen (n = 16) and air sac (n = 16)] at Ismailia Governorate, Egypt. Average broiler chicken age was 7 weeks and the average weight was 1.8 kg. Handling of birds was performed according to the Animal Ethics Review Committee of Suez Canal University, Egypt. Samples were collected in the period from November 2016 until August 2017. The collected samples were prepared for bacteriological examination.
Isolation and identification of E. coli
The collected specimens were inoculated in peptone water and then incubated at 37 °C for 24 h. A loopful from the incubated broth was streaked onto MacConkey′s agar and EMB plates and then incubated at 37 °C for 24 h. Suspected colonies were identified by microscopical examination, cultural characters as well as biochemical reactions as described by Quinn et al. (2011).
Serotyping of E. coli strains
The isolates of E. coli were subjected to serotyping where somatic (O) antigen was investigated by slide agglutination test as described by Edwars and William (1972). Flagellar (H) antigen stereotyping was performed as described by Davies and Wray (1997).
Antimicrobial susceptibility testing
Escherichia coli strains were tested against 12 antimicrobial agents (ampicillin, amoxicillin/clavulanic acid, erythromycin, gentamicin, neomycin, tetracycline, doxycycline, levofloxacin, norfloxacin, trimethoprim/sulphamethoxazole, sulphamethoxazole, and colistin sulphate) according to the methods described by NCCLS (2015) using disc diffusion technique. The susceptibility was determined according to the size of the inhibition zone. Multidrug resistance (MDR) was categorized for resistance to two or more unique antibiotic classes.
PCR detection of virulence and disinfectant resistance genes of E. coli
Escherichia coli serotypes (n = 113; 23 environmental strains and 90 strains of avian origin) were tested for the detection of 4 virulence genes (iss, papC, eaeA, and cfaI) and 3 QACs resistance genes (qacEΔ1, QacA/B, and QacC/D) by using PCR. The DNA extraction was performed as described by the instructions of QIAamp DNA mini kit. The reaction volume includes (6 μl of the extracted DNA, 12.5 μl of Emerald Amp GT PCR master mix (2× premix) and 1 μl of each primer forward and reverse, PCR grade water 4.5 μl). Positive control strains were kindly given by Animal Health Research Institute, Dokki, Egypt. Primers used in PCR were illustrated in Table 1. PCR Protocol: initial denaturation at 94 °C for 5 min; denaturation at 94 °C for 30 s; annealing at 54 °C for 30 s for iss gene, at 51 °C for 30 s for eaeA, at 58 °C for 40 s for papC, at 50 °C for 40 s for cfaI, 58 °C for 40 s for qacEΔ1 gene, at 53 °C for 40 s for qacA/B and at 53 °C for 30 s for qacC/; extension at 72 °C for 30 s in iss, eaeA and qacC/D, at 72 °C for 40 s in papC and cfaI qacEΔ1 and qacA/B; cycles repeated for 35 times. Finally, the PCR products were separated by using electrophoresis and then photographed.
Statistical analyses
The data frequencies were analyzed by the nonparametric test (Chi square) with the aid of SAS (2004) software to test the null hypothesis of different treatment groups. The level of significance was P < 0.05.
Results
Prevalence of E. coli in diseased broiler chickens and farm environment
The bacteriological examination of 142 environmental samples revealed 38 E. coli strains (26.76%) including; feeders, drinkers, walls and floors samples with percentages of 37.5%, 31.25%, 11.11%, 28.57%, respectively. While 226 organ samples, revealed 114 E. coli strains with a prevalence of (50.44%). E. coli was isolated from internal organs (heart, liver, lung, yolk, spleen, air sac) with percentages of 42.86%, 60.98%, 33.33%, 20%, 50%, 100%, respectively. The total E. coli prevalence was (41.30%) as illustrated in Table 2.
Serotyping of the isolated E. coli strains
In this study, 38 E. coli strains originated from environmental samples were subjected to serological identification, 23 strains (60.5%) have belonged to the following 10 different serotypes: O78 (13.16%), O119:H4, O113:H4, O169, O91:H21, O142, O111:H2, O1:H7, O26:H11 and O128:H2 (5.26% for each), while 15 strains (39.5%) were untypable. In addition, 114 E. coli strains (originated from organs of diseased broilers) were subjected to serological identification, 90 strains (78.95%) have belonged to the following 12 different serotypes: O1:H7 (13.16%), O78 (13.16%), O126 (8.77%), O91:H21 (8.77%), O125:H21, O44:H18, O121:H7, O15:H2, O146:H21, O124, O20 and O128:H2 (4.39% for each), moreover 24 strains (21.05%) were untypable as described in Table 3.
Antimicrobial susceptibility of E. coli strains
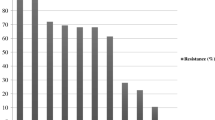
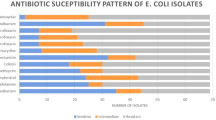
As described in Table 4, the antimicrobial susceptibility testing of the isolated strains proved that, the tested strains were highly resistant (100%) to, ampicillin, erythromycin and tetracycline, followed by amoxicillin-clavulanic acid, norfloxacin, and sulphamethoxazole (80.92% for each), trimethoprim/sulphamethoxazole (75%) and gentamycin (50%). While (100%) of the tested strains were sensitive to colistin sulphate, followed by neomycin (87.5%). Meanwhile, the tested strains were intermediate sensitive to doxycycline (75%) and levofloxacin (62.5%).
PCR detection of virulence genes and QACs resistance genes
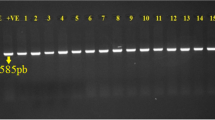
PCR was used for detection and amplification of 4 virulence genes (iss, papC, eaeA and CFAI) in the isolated strains as illustrated in Table 5, where (100%) of the tested strains were positive for iss gene at specific amplicon size 266 bp (Fig. 1a) and papC gene with specific amplicon size 501 bp (Fig. 1b). Only (20.3%) of the tested isolates were positive for eaeA gene with specified amplicon size 248 bp (Fig. 1c), the positive strains including; O1:H7 (n = 12), O78 (n = 5), O128:H2 (n = 2), O119:H4 (n = 2) and O113:H4 (n = 2). In addition, cfaI gene was absent in all examined strains as shown in (Fig. 1d).
Electrophoretic pattern of PCR products of E. coli iss, papC, eaeA, and cfaI genes: a (L): the DNA molecular size ladder (Gelpilot 100 bp ladder); (Neg.): negative control; (Pos.): positive control; (lanes 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10): positive amplification of 266 bp of iss gene of different E. coli strains. b (L): the DNA molecular size ladder (Gelpilot 100 bp ladder); (Neg.): negative control; (Pos.): Positive control; (lanes 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10): positive amplification of 501 bp of papC gene of different E. coli strains. c (L): the DNA molecular size ladder (Gelpilot 100 bp ladder); (Neg.): negative control; (Pos.): Positive control; (lanes 1 and 6): positive amplification of 248 bp of eaeA gene; (lanes 3,4,5,7,8,9,10): Negative strains. d (L): the DNA molecular size ladder (Gelpilot 100 bp ladder); (Neg.): negative control; (Pos.): positive control; (lanes 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10): negative amplification of 364 bp of cfaI gene of different E. coli strains
Concerning the detection of the QACs resistance genes, PCR was used for the detection and amplification of (qacAΔ1, qacA/B and qacC/D) genes in the isolated strains, all the tested strains (100%) were positive for qacAΔ1, qacA/B and qacC/D genes with specific amplicon size 362 bp, 361 bp and 195 bp, respectively, as illustrated in Fig. 2a–c and Table 5.
Electrophoretic pattern of PCR products of E. coli qacEΔ1, QacA/B, and QacC/D genes: a (L): the DNA molecular size ladder (Gelpilot 100 bp ladder); (Neg.): negative control; (Pos.): positive control; (lanes 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10): positive amplification of 362 bp of qacEΔ1 gene of different E. coli strains. b (L): the DNA molecular size ladder (Gelpilot 100 bp ladder); (Neg.): negative control; (Pos.): positive control; (lanes 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10): positive amplification of 361 bp of qacA/B gene of different E. coli strains. c (L): the DNA molecular size ladder (Gelpilot 100 bp ladder); (Neg.): negative control; (Pos.): positive control; (lanes 1, 2, 3, 4, 5, 6, 7, 8, 9 and 10): positive amplification of 195 bp of qacC/D gene of different E. coli strains
Discussion
Cross-resistance between antibiotics and QAC could occur by various mechanisms on the same resistance plasmid and or transposon (Hegstad et al. 2010). The presence of Quaternary Ammonium Compounds determinants on various mobile constituents helps in the transport of resistance to another microorganism (Gillings et al. 2009). The massive and improper application of antibiotics for long-term in poultry farms resulted in multidrug resistance in different bacterial pathogens (Singer and Hofacre 2006). In the present work, the bacteriological examination showed that the prevalence of E. coli in environmental specimens was (26.76%), while was (50.44%) in diseased broiler chickens. The total prevalence of E. coli was (41.30%) as illustrated in Table 2. The prevalence of E. coli from different environmental sources is not significantly differed (P > 0.05). Meanwhile, higher significant differences were recorded (P < 0.0001) in the prevalence of E. coli from organs of diseased broiler chickens compared to those of the environmental sources. The highest prevalence was recorded in air sac (100%) followed by liver (60.98%), while the lowest prevalence was recorded in the yolk (20%). Higher prevalence of (84%) was obtained by Oboegbulem et al. (2009) who isolated this organism from commercial and backyard poultry farms and chicken markets. Multiple predisposing conditions could rise the susceptibility of poultry to colibacillosis, including; respiratory viruses, overcrowding, bad handling of birds and bad sanitation (Eid et al. 2016).
Regarding the serotyping of the isolated E. coli strains, 38 E. coli strains isolated from environmental samples were subjected to the serological identification, 23 strains (60.5%) have belonged to 10 serogroups and the most predominant serogroup was O78 (13.16%), while 15 E. coli strains (39.5%) were untypable. In addition, 114 E. coli strains originated from organs of diseased broilers were subjected to serological identification, 90 strains (78.95%) have belonged to 12 different serogroups and the most predominant serogroups were O1:H7 (13.16%), O78 (13.16%), O126 (8.77%), O91:H21 (8.77%), while 24 strains (21.05%) were untypable, as described in Table 3. There is a significant difference in the prevalence of the isolated serotypes (P < 0.0243). These results disagree with those obtained by Yousef et al. (2015) who recorded that the most prevalent serotypes originated from different sources of poultry broiler farms were untypable E. coli serovars; followed by O26; then O2, O124, O125, and O114. Chart et al. (2000) proved that the Avian Pathogenic E. coli are mainly included specific serotypes, especially serotypes O78, O2 and O1, followed by O55, and O15. The emergence of certain serotype and its responsibility for disease occurrence is mainly depending upon the health condition of chicken, the environmental conditions, handling and management procedures (Srinivasan et al. 2013).
In the present study, the isolated E. coli strains were tested against 12 antimicrobial agents, The resistance and sensitivity of the isolated strains to different antimicrobial discs were differed significantly (P < 0.0001) as described in Table 4. The tested strains showed multiple drug resistance and were highly resistant (100%) to ampicillin, erythromycin and tetracycline, followed by amoxicillin/clavulanic acid, norfloxacin, and sulphamethoxazole (80.92%), trimethoprim/sulphamethoxazole (75%) and gentamycin (50%). While (100%) of the tested strains were sensitive to colistin sulphate, followed by neomycin (87.5%). These results are nearly agreed with those obtained by Hashem et al. (2012) and Ola (2017).
Production of β -lactamase enzyme that breaks down the beta-lactam ring of penicillin is the major mechanism of antibiotic resistance in E. coli. Gene encoding β -lactamase enzyme could be carried on plasmid or on bacterial chromosome (Udomsantisuk et al. 2011), while aminoglycosides resistance is mainly taking place in pathogenic E. coli due to aminoglycoside modifying enzyme (Galimand et al. 2003) which is encoded on R-plasmids (González-Zorn et al. 2005). Sulfonamides, penicillins and tetracyclines, and are the most popular and the oldest antimicrobial agents that used heavy against bacterial infection so that a high level of drug resistance has emerged with the time (Joint 2008).
Regarding the genetic detection of virulence genes, in the present study PCR was used for detection and amplification of (iss, papC, eaeA, and cfaI) genes, as illustrated in Table 5 (100%) of the tested strains were positive for iss gene at specific amplicon size 266 bp (Fig. 1a) and papC gene with specific amplicon size 501 bp (Fig. 1b). Only (20.3%) of the tested strains were positive for eaeA gene with specified amplicon size 248 bp (Fig. 1c), furthermore, all the examined strains were negative for cfaI gene as illustrated in (Fig. 1d). These results are agreed with those described by Dissanayake et al. (2014) who reported that 85.4% of Avian Pathogenic E. coli which originated from diseased birds suffering from colibacillosis in the USA were positive for iss gene, also this result is supported by Ewers et al. (2004) who mentioned that iss gene was detected in 82.7% of APEC strains originated from birds suffering from coli septicemia in Germany. In contrary to the results of papC gene in this study Rodriguez-Siek et al. (2005) stated that the papC gene is commonly associated APEC with a percentage (44.1%).
Moreover, attaching and effacing is a term that used to clarify the lesion caused by E. coli in the host intestine, where (attaching) refers to the intimate adhesion to the cell membrane of intestinal cells, while (effacing) refers to the destruction of intestinal microvilli (Stordeur et al. 2000). In this study, the prevalence of eaeA gene was (20.3%), these results are agreed with Ola (2017) who reported that the incidence rate of eaeA gene in the tested E. coli strains was 15.79%. Also, these results nearly agreed with those obtained by (DebRoy and Maddox 2001). In contrary to these findings, Ramadan et al. (2016) stated that all the tested E. coli strains derived from chicken viscera were carried the eae gene (100%).
Concerning the detection of the disinfectant resistance genes, PCR was used for detection and amplification of QACs resistance (qacAΔ1, qacA/B, qacC/D) genes in the isolated E. coli strains, as illustrated in Table 5 all the tested E. coli strains (100%) were positive for qacAΔ1, qacA/B and qacC/D genes with specific amplicon size 362 bp, 361 bp and 195 bp, respectively, as illustrated in (Fig. 2a–c). These results are supported by Amira (2016) who found that the distribution of qacEΔ1 was (93.1%). The massive use of QACs in the farm environment could result in acquired QACs resistance in E. coli strains (Sidhu et al. 2002). Many QACs resistant genes are commonly associated with multidrug-resistant pathotypes especially qacC/D, qacA/B, and qacE (Zhang et al. 2015). The qacEΔ1gene is common in enteric bacterial pathogens possessing Sulphonamide resistant determinants. Seventy percent of qacEΔ1- +ve strains exhibit cross-resistance to Sulphamethoxazole, 60% of qacEΔ1- +ve strains exhibits cross-resistance to Sulfamethoxazole-trimethoprim. Also, 40% of qacEΔ1- +ve strains were highly resistant to Gentamicin (Kücken et al. 2000).
In conclusion, E. coli continues to be one of the most important pathogens in poultry and poultry farm environment, the most predominant E. coli serotypes affecting broiler chickens are O78, O1:H7, O91:H21 and O126. The QACs resistance genes are frequently distributed with the multidrug-resistance pathotypes which may be transmitted to humans by the consumption of chickens or any byproduct containing such strains. The high proportion of virulence genes (iss and papC) and the multidrug-resistance phenomena is prevalent in Avian Pathogenic E. coli and environmental strains. There is a directly proportional relationship between the presence of multidrug-resistance, disinfectant resistant genes, virulence genes and the severity of lesions associated with E. coli infection and complications.
Availability of data and materials
Not applicable.
References
Amira FA (2016) Molecular characterization of virulence genes in Salmonella spp. isolated from poultry. Ph.D. Kafrelshikh University, Egypt
Barnes HJ, Nolan LK, Vaillancourt JF (2008) Colibacilliosis. In: Saif AM (ed) Diseases of poultry. Blackwell Publishing, New York, pp 691–732
Bisi-Johnson MA, Obi CL, Vasaikar SD, Baba KA, Hattori T (2011) Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog 3:9. https://doi.org/10.1186/1757-4749-3-9
Bore E, Hebraud M, Chafsey I, Chambon C, Skjæret C, Moen B, Møretrø T, Rudi K, Langsrud S (2007) Adapted tolerance to benzalkonium chloride in Escherichia coli K-12 studied by transcriptome and proteome analyses. Microbiology 153:935–946
Buffet-Bataillon S, Le Jeune A, Le Gall-David S, Bonnaure-Mallet M, Jolivet-Gougeon A (2012) Molecular mechanisms of higher MICs of antibiotics and quaternary ammonium compounds for Escherichia coli isolated from bacteraemia. J Antimicrob Chemother 67:2837–2842
Chart H, Smith H, La Ragione R, Woodward MJ (2000) An investigation into the pathogenic properties of Escherichia coli strains BLR, BL21, DH5α and EQ1. J Appl Microbiol 89:1048–1058
Chuanchuen R, Khemtong S, Padungtod P (2007) Occurrence of qacE/qacED1 genes and their correlation with class 1 integrons in Salmonella enterica isolates from poultry and swine. Southeast Asian J Trop Med Public Health 38:855–862
Correa J, De Paulis A, Predari S, Sordelli D, Jeric P (2008) First report of qacG, qacH and qacJ genes in Staphylococcus haemolyticus human clinical isolates. J Antimicrob Chemother 62:956–960
Davies R, Wray C (1997) Immunomagnetic separation for enhanced flagellar antigen phase inversion in Salmonella. Lett Appl Microbiol 24:217–220
De Carli SN, Ikuta FKM, Lehmann VP, da Silveira G, de Melo Predebon ASK, Lunge VR (2015) Virulence gene content in Escherichia coli isolates from poultry flocks with clinical signs of colibacillosis in Brazil. Poult Sci 94:2635–2640
DebRoy C, Maddox CW (2001) Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim Health Res Rev 2:129–140
Dissanayake D, Octavia S, Lan R (2014) Population structure and virulence content of avian pathogenic Escherichia coli isolated from outbreaks in Sri Lanka. Vet Microbiol 168:403–412
Edwars PR, William HE (1972) Identification of Enterobacteriaceae, 3rd edn. Burges Puplication Company, Burges
Eid HI, Algammal AM, Nasef SA, Elfeil WK, Mansour GH (2016) Genetic variation among avian pathogenic E coli strains isolated from broiler chickens. Asian J Anim Vet Adv 11:350–356. https://doi.org/10.3923/ajava.2016.350.356
Eid HM, Algammal AM, Elfeil WK, Youssef FM, Harb SM, Abd-Allah EM (2019) Prevalence, molecular typing, and antimicrobial resistance of bacterial pathogens isolated from ducks. Vet World 12(5):677–683
Ewers C, Janßen T, Kießling S, Philipp HC, Wieler LH (2004) Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet Microbiol 104:91–101
Fraise AP, Maillard J, Sattar S (2013) Principles and practice of disinfection, preservation and sterilization. Wiley, Hoboken
Galimand M, Courvalin P, Lambert T (2003) Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother 47:2565–2571
Ghosal A, Bhowmick R, Nandy RK, Ramamurthy T, Chatterjee NS (2007) PCR-based identification of common colonization factor antigens of enterotoxigenic Escherichia coli. J Clin Microbiol 45:3068–3071
Gillings MR, Holley MP, Stokes H (2009) Evidence for dynamic exchange of qac gene cassettes between class 1 integrons and other integrons in freshwater biofilms. FEMS Microbiol Lett 296:282–288
González-Zorn B, Teshager T, Casas M, Porrero MC, Moreno MA, Courvalin P, Domínguez L (2005) armAand aminoglycoside resistance in Escherichia coli. Emerg Infect Dis 11:954–956. https://doi.org/10.3201/eid1106.040553
Hashem M, Elahi M, Mannan M, Kabir M, Kashem M, Pallab M (2012) Isolation, identification and antibiogram of Escherichia coli from broiler at Chittagong district in Bangladesh. Wayamba J Anim Sci 4:312–316
Hegstad K, Langsrud S, Lunestad BT, Scheie AA, Sunde M, Yazdankhah SP (2010) Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health? Microbial Drug Resistance 16:91–104
Jin W, Zheng Z, Zhang Y, Qin A, Shao H, Liu Y, Wang J, Wang Q (2008) Distribution of virulence-associated genes of avian pathogenic Escherichia coli isolates in China. Agric Sci China 7:1511–1515
Joint (2008) FAO/WHO/OIE Expert Meeting on Critically Important Antimicrobials: report of the FAO/WHO/OIE Expert Meeting, FAO Headquarters, Rome, 26–30 November. Food and Agriculture Organization of the United Nations, Rome
Kücken D, Feucht H, Kaulfers P (2000) Association of qacE and qacE Δ1 with multiple resistance to antibiotics and antiseptics in clinical isolates of Gram-negative bacteria. FEMS Microbiol Lett 183:95–98
Mellata M (2013) Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Path Dis 10:916–932
NCCLS (2015) Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement according to CLSI. CLSI document M100-s15. In: CLSI document M100-s15. N. C. F. C. L. standard ed. Clinical Laboratory standard Institute, Wayne
Noguchi N, Suwa J, Narui K, Sasatsu M, Ito T, Hiramatsu K, Song JH (2005) Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J Med Microbiol 54:557–565
Oboegbulem S, Abiade C, Onunkwo J, Ezenduka E, Chah F, Nwanta J, Anosike C (2009) Incidence of verotoxigenic Escherichia coli in poultry in Nsukka urban area of southeastern Nigeria. Anim Sci Report 3:128–131
Ola AI (2017) Bacteriological and molecular studies on bacteria causing omphalitis in chicks. Ph.D. Benha University, Egypt
Quinn P, Bryan K, Finola C, Hartigan P, Fitzpartrick ES (2011) Veterinary microbiology and microbial diseases. Wiley, New York
Ramadan H, Awad A, Ateya A (2016) Detection of phenotypes, virulence genes and phylotypes of avian pathogenic and human diarrheagenic Escherichia coli in Egypt. J Infect Develop Ctries 10:584–591
Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Nolan LK (2005) Characterizing the APEC pathotype. Vet Res 36(2):241–256
SAS (2004) Statistical user’s Guide. INT., Cary, NC. USA
Sidhu MS, Heir E, Sørum H, Holck A (2001) Genetic linkage between resistance to quaternary ammonium compounds and β-lactam antibiotics in food-related Staphylococcus spp. Microbial Drug Resistance 7:363–371
Sidhu MS, Sørum H, Holck A (2002) Resistance to quaternary ammonium compounds in food-related bacteria. Microbial Drug Resistance 8:393–399
Singer RS, Hofacre CL (2006) Potential impacts of antibiotic use in poultry production. Avian Dis 50:161–172
Srinivasan P, Balasubramaniam GA, Murthy GK, Balachandran P (2013) Bacteriological and pathological studies of egg peritonitis in commercial layer chicken in Namakkal area. Asian Pac J Trop Biomed 3:988–994
Stordeur P, China B, Charlier G, Roels S, Mainil J (2000) Clinical signs, reproduction of attaching/effacing lesions, and enterocyte invasion after oral inoculation of an O118 enterohaemorrhagic Escherichia coli in neonatal calves. Microbes Infect 2:17–24
Udomsantisuk NP, Nunthapisud T, Tirawatanapong T, Dansuputra M (2011) Molecular characterization of extended spectrum beta-lactamase among clinical isolates Escherichia coli and Klebsiella pneumoniae. J Med Assoc Thai 94:1504–1512
Yaguchi K, Ogitani T, Osawa R, Kawano M, Kokumai N, Kaneshige T, Noro T, Masubuchi K, Shimizu Y (2007) Virulence factors of avian pathogenic Escherichia coli strains isolated from chickens with Colisepticemia in Japan. Avian Dis 51:656–662. https://doi.org/10.1637/0005-2086(2007)51%5b656:vfoape%5d2.0.co;2
Yousef SA, Ammar AM, Ahmed DA (2015) Serological and molecular typing of avian pathogenic E. coli originating from outbreaks of colibacillosis in chicken flocks. Int J Sci Res 4:2082–2088
Zhang C, Cui F, Zeng GM, Jiang Z, Yang Z, Yu M, Zhu Y, Shen L (2015) Quaternary ammonium compounds (QACs): a review on occurrence, fate and toxicity in the environment. Sci Total Environ 518:352–362
Acknowledgements
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.
Author information
Authors and Affiliations
Contributions
MEE and AMA conceived and designed the experiments and performed the experiments. AMA, SAN, SAMA and MB analyzed the data. AMA, MEE, AET and AAA wrote and revised the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Handling of birds was performed according to the Animal Ethics Review Committee of Suez Canal University, Egypt.
Consent for publication
All authors gave their informed consent prior to their inclusion in the study.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Enany, M.E., Algammal, A.M., Nasef, S.A. et al. The occurrence of the multidrug resistance (MDR) and the prevalence of virulence genes and QACs resistance genes in E. coli isolated from environmental and avian sources. AMB Expr 9, 192 (2019). https://doi.org/10.1186/s13568-019-0920-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-019-0920-4