Abstract
Background
Apart from localized gastrointestinal infections, Escherichia coli and Salmonella species are major causes of systemic disease in both humans and animals. Salmonella spp. cause invasive infections such as enteric fever, septicemia, osteomyelitis and meningitis while certain types of E. coli can cause systemic infections, including
pyelonephritis, meningitis and septicemia. These characteristic requires the involvement of a myriad of virulence factors.
Methods
This study investigated the virulence factors of Escherichia coli and Salmonella species in clinical specimens from patients with diarrhoea presenting to health care centres in Oliver R. Tambo District Municipality, Eastern Cape Province, Republic of South Africa. Microbiology analysis involved the use of cultural and molecular techniques.
Results
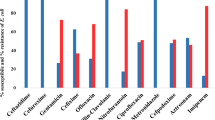
Out of a total of 315 samples screened, Salmonella isolates were obtained in 119 (37.8%) of cases and these comprised: S. choleraesuis (6%), S. enteritidis (4%), S. eppendorf (1%), S. hadar (1%), S. isangi (8%), S. panama (1%), S. typhi (52%), S. typhimurium (25%) and untyped Salmonella spp. (2%). Among the Salmonella species 87 (73.1%) were invasive. Using molecular diagnostic methods, diarrheagenic E. coli were detected in 90 cases (28.6%): the greater proportion of this were enteroaggregative E. coli (EAEC) 37 (41.1%), enteropathogenic E. coli (EPEC) 21 (23.3%) and enterohemorrhagic E. coli (EHEC) 21 (23.3%). The predominant virulence gene among the diarrheagenic E. coli was EAEC heat-stable enterotoxin astA genes while the virulence genes identified in the Salmonella strains were 15 (12.6%) flic and 105 (88.2%) inv genes. The amino acid identity of the representative genes showed 95-100% similarity to corresponding blast searched sequence.
Conclusions
This study showed the diversity of virulence gene expression in two major enteric pathogens. S. typhi and enteroaggregative E. coli were the predominant enteropathogens in our study area with an indication that EAEC is endemic within our study population. It was observed among other things that some diarrheagenic E. coli isolated from apparently asymptomatic subjects expressed some virulence genes at frequency as high as seen in diarrheagenic cases. This study underlines the importance of understanding the virulence composition and diversity of pathogens for enhanced clinico-epidemiological monitoring and health care delivery.
Similar content being viewed by others
Background
Gastrointestinal infections due to pathogenic Enterobacteriaceae in particular Escherichia and Salmonella species are significant causes of morbidity and mortality worldwide. These infections which usually are self-limiting may be fatal in hosts with debilitating immune systems [1]. The fatality of infections due to these enteric pathogens depends on their serotypes, the size of the inoculum, and the status of the host [2]. Escherichia and Salmonella species were reported to have diverged from a common ancestor based on the evolutionary rate estimates from 5S and 16S rRNA sequence analyses while Shigella spp. are considered clonal lineages of Escherichia coli[3]. Salmonella species are mainly pathogenic, with differing host ranges. S. typhi is adapted to humans and does not occur in animals while non-typhoidal Salmonella serovars (NTS) have a broad vertebrate host range [2]. Even though E. coli is generally known as commensal normal flora of the gut, some E. coli strains are the causative agents of neonatal meningitis, urinary tract infections, bacteremia, and infectious diarrhea.
The major distinguishing factor between pathogenic and non-pathogenic strains of E. coli strains is the occurrence of virulence genes, which code for the various known strategies for pathogenecity. Analysis have shown that pathogenic E. coli strains from diarrhoea cases and those involved in urinary tract infections are more of a distinct subsets of E. coli, rather than a reflection of the random fecal flora [4]. Some of the virulence factors of E. coli include ability to adhere, colonize, and invade the hosts' cells. Further to these are the secretion systems, production of cell surface molecules, transport and siderophore formation [5]. According to Kaper et al., [6], E. coli has been categorized based on the type of virulence factors present and host clinical symptoms basically into the following pathotypes: enteropathogenic E. coli (EPEC); enterohemorrhagic E. coli (EHEC); enterotoxigenic E. coli (ETEC); enteroaggregative E. coli (EAEC) and diffusely adherent E. coli (DAEC), a subclass of enteroaggregative E. coli; enteroinvasive E. coli (EIEC); uropathogenic E. coli (UPEC) and neonatal meningitis E. coli (NMEC).
The ability of the enteric pathogen to invade and penetrate intestinal epithelial cells is required in salmonellosis whether it is confined as the intestinal form or progresses to systemic involvement [7]. The attribute to direct their internalization by the epithelial cells which are not normally phagocytic is a striking Salmonella-host cell interaction. According to Galan and Curtiss [8] this remarkable phenotype known as invasion allowed for identification and characterization of invasion genes. The key mechanism involves type III secretion systems which are encoded by pathogenicity island 1 (SPI-1) [9]. Salmonella also possess the ability to alter phagocytosis in order to circumvent the process. S. enterica serovar Typhimurium is known to delay significantly the fusion of the phagosome to the lysosome [10]; thereby hibernating in phagocytic cells and hence adapt to resist the antimicrobial activity of the fused phagolysosome [11]. Bacterial survival in phagocytic cells has been observed as an alternate to invasion in accessing privileged sites in hosts. Rescigno et al., [12] postulated that CD18+ expressing phagocytes are alternate route and these cells have been observed by Vasquez-Torres et al., [13] as vehicles for reaching the spleen in an invasion-independent manner by S. enterica serovar Typhimurium.
Molecular analysis is known to give a better picture of epidemiology of infectious diseases. Studies have demonstrated the sensitivities of molecular-based methods to be greater compared to current conventional methods of analysis [1]. Molecular studies have led to the understanding of the genomic make-up of bacteria which generally consist of stable regions and variable regions, the flexible part that is composed of bacteriophages, plasmids, transposons as well as unstable large regions, called genomic islands. The so-called genomic islands are a gene pool required for encoding virulence factors of pathogenic bacteria and these have been designated "pathogenicity islands" [14]. The concept of pathogenicity islands (Pais) was first identified through the genetic and molecular analysis of virulence genes in uropathogenic E. coli and EPEC [15]. Pais which are specific regions of chromosomal DNA have been described in more than 30 bacterial species [14]. It is a well known concept that bacterial pathogenicity is an organized multifactorial process involving numerous chromosomal and extrachromosomal genes directed by complex regulatory circuits [16, 17].
There are various shared genetic strategies for pathogenicity in enteric bacilli. Type III secretion is a dedicated secretion machinery whose components are coded for by numerous homologous gene sequences shared by enteric pathogens [3, 18]. Nevertheless, there has been understanding that the similarities between EPEC virulence attributes and Salmonella invasion genes are more than homologous genes associated with secretion [19]. Most virulence factors of pathogenic E. coli, Shigella, and Salmonella strains are plasmid-borne however; one or more of the essential virulence determinants are borne on an extrachromosomal element [20]. In both E. coli and Salmonella spp. fimbriae might play a role in adhesion and invasion [21]. The curli fimbriae of these strains were proven to bind to several tissue-matrix proteins as well as plasminogen and its activator t-PA [3].
Bacteria are emerging with new means of circumventing human efforts at curbing their nefarious schemes and various evolvement patterns and innovations are certainly put in place by these pathogens. A myriad combination of virulence genes against indiscriminate genetic transfer and recombination are required for a successful emergence of pathogen [22, 23]. Profiling the expression of these genes will give impetus to understanding the mechanisms by which enteric bacterial pathogens colonize, spread and at times persist in the hosts [24]. This study investigated the genetic determinants of virulence in E. coli and Salmonella spp. which are significant pathogens involved in enteric diseases.
Methods
Specimens' collection and bacterial isolation
The study is a retrospective and cross-sectional study which utilized data originally collected for surveillance purposes focusing on Nelson Mandela Academic Hospital Complex (NAMHC), Mthatha, a health facility which serve as a referral hospital in the Eastern Cape Province of South Africa. Specimens' collection from cases was from both male and female diarrhoeic patients from all age categories and was based on availability. Written informed consent was obtained from all patients, parents or guardians as the case may be and questionnaire was administered by trained volunteer health workers. Information provided include the frequency of episodes of diarrhoea and whether or not antibiotic or other forms of medication has been used. The control specimens were then selected from the group which had not had diarrhea or antibiotic therapy in the preceding 2 weeks. The study protocol and data handling were approved by the WSU ethics committee (Protocol No. 0003/08) as well as the Department of Health, Eastern Cape, South Africa.
Salmonella isolates deposited at the NICD, Johannesburg under the surveillance study of 2005 to 2008 from this tertiary health facility were obtained. For 2009 fresh stool samples in sterile stool jar or rectal swabs in Cary-Blair transport medium per patient were collected from 125 patients presenting with acute diarrhoea in the tertiary referral facility and surrounding clinics and 75 apparently healthy school pupils in three different schools within ORTDM. These were transported on ice pack to the laboratory where analysis was done within 24 hours. Where this is not possible specimen were preserved at temperature between 4 to 8°C.
Bacteriological analyses
NICD isolates
Bacteriological analyses of the specimens for these isolates were carried out at the National Health Laboratory Services, Nelson Mandela Academic Hospital, Mthatha. Samples were examined for the presence of E. coli, Salmonella and Shigella using standard conventional methods according to Forbes et al., [25].
E. coli
The faecal samples were cultivated on MacConkey agar. After overnight incubation at 37°C, lactose fermenting colonies (LFC) with the typical appearance of E. coli were selected for further analysis. Isolates were identified by biochemical assays using Microscan Gram negative combo panel NUC 45 (Siemens/Dade Behring).
Salmonella and Shigella
Specimens were cultivated for the isolation of Salmonella and Shigella species on MacConkey agar. After 24 h of incubation at 37°C, suspected colonies with typical characteristics of Salmonella and Shigella were sub-cultured on XLD (xylose lysine deoxycholate) agar for 24 h at 37°C. Confirmation was carried out using Microscan Gram negative combo panel NUC 45 (Siemens/Dade Behring).
DNA extraction
DNA template for PCR was obtained from pure overnight bacterial culture using Fungal/Bacterial DNA extraction kit™ (Zymo Research) and following manufacturer's instructions. The concentration of the eluted DNA was measured using NanoDrop 2000 spectrophotometer (Thermo Scientific).
DNA amplification
PCR amplifications were performed in a final volume of 25 μℓ containing: 0.5 to 2 μℓ of DNA template depending on concentration, 8.5 to 10 μℓ of Nuclease free water, 1 μℓ of each primer and 12.5 μℓ Master mix (EconoTaq Green, Fermentas). Amplifications were carried out in a GeneAmp PCR System 9700 Thermocycler (Applied Biosystems). All oligonucleotide primers were synthesized by Inqaba Biotechnology (Pretoria, South Africa) and the sequences are as shown in Table 1 (63-72). The PCR cycling conditions for the E. coli strains consisted of 95°C for 5 min while for the Salmonella isolates consisted of 95°C for 1 min, which were followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 30 s. Amplification products were separated by electrophoreses on 10 mg/ml agarose gel (TopVision TM, Fermentas) in 1× TBE Buffer and ethidium bromide (5 μℓ) with a 100-bp ladder (Fermentas) as molecular weight marker.
Sequencing reaction
PCR products were sequenced using an Applied Biosystems 3500xL Genetic analyzer (AB Biosystems). Prior to PCR products sequencing, the unincorporated dNTPs were dephosphorylated with a commercial kit from Zymo Research Corporation (Orange, CA). Subsequently, the PCR products were sequenced with the ABI PRISM BigDye terminator cycle sequencing ready reaction kit (AB Biosystems) using the same primers as employed in the PCR reactions. The products were then subjected to the following conditions: 94°C for 2 min, followed by 40 cycles of denaturation at 85°C for 1 s, annealing at 53°C for 10 s and extension at 60°C for 2 min 30 s, with a final extension at 4°C for 0 s. The sequencing reaction products were cleaned up using ZR-96 DNA sequencing clean-up kit™. Thereafter, the ultra-pure products were analyzed on the sequencing machine. Sequences were aligned with known E. coli and Salmonella virulence gene sequences by a blast search of the National Center for Biotechnology Information (NCBI) data base http://www.ncbi.nlm.nih.gov/BLAST/ using Staden package version 1.6.0-beta4 (MRC.WTSI).
Results
Demographic features
Patients' data were analyzed using Microsoft Excel version 2003. Continuous variables were summarized as mean. Our subjects were predominated by male patients 165/315 (52.4%) and no age group was excluded, with the youngest patient being 3 months and the oldest 91 years. A sizeable number of cases, 95 (30.15%) were between the ages 7 to 13 and controls school-aged 7 to 12 were matched for this age category.
Bacteriological identification and Molecular analysis
Results showed that Salmonella strains were isolated from 119 (37.8%) of cases while diarrheagenic E. coli was found in 90 (28.6%) of cases. The distributions of the different pathotypes are as shown in Figures 1 and 2. Of the Salmonella isolates 87 (74.1%) were invasive. The most common virulence factors detected among the Salmonella strains were invA found in 105 Salmonella spp. and fliC genes detected in 15 Salmonella isolates. The predominant virulence gene among the diarrheagenic E. coli was 24 EAEC heat-stable enterotoxin astA genes. Table 2 showed the distribution of the various genes among cases and controls. The representative gels for PCR amplification of DNA extracted from selected E. coli and Salmonella isolates showing the presence of diverse virulence genes are indicated in additional files 1A, B and 2 respectively. One hundred and eighty isolates were obtained from the 150 control subjects. E. coli was the predominant bacterial species being 85 (47.2%) while Salmonella spp. was 8 (12.1%). Other recovered bacteria species were Proteus mirabilis 45 (25.0%), Klebsiella pneumoniae 23 (12.8%) and Enterobacter cloacae 19 (10.5%). The sequencing analysis of our genes showed 100% conformation of the various virulence genes with corresponding blast search sequence and confirmed the strain.
Discussion
Gastroenteritis is a major concern in sub-Saharan Africa as with other developing countries [26]. South African National Burden of Disease study of the year 2000 found that diarrhoea accounted for nearly 3% of all deaths in South Africa [27]. According to the South African health review of 2007, death due to gastroenteritis among children was put at 15% [28] showing increasing mortality. The developed countries are not spared in the global burden of enteric-related diarrhoea. Salmonellosis was considered a major public health problem in the United States [29]. E. coli and Salmonella are among the bacterial pathogens implicated in gastroenteritis. These enteric pathogens have evolved different strategies for subverting normal host cellular functions [30]. These pathogens cause various intestinal and extraintestinal diseases by means of virulence factors that affect a wide range of cellular processes. These virulence induced infections usually involve complex mechanisms with various specific, interdependent interactions between hosts and pathogens [31]. This present study provides information on the pathotypes and some virulence factors associated with local isolates of E. coli and Salmonella species. E coli are more than just a harmless intestinal microflora; it can also be a highly versatile, and frequently deadly, pathogen [6]. E. coli strains cause diarrhoea by several distinct pathogenic mechanisms and differ in their epidemiology. Virulence genes were expressed in diarrheagenic E. coli from both cases and controls. EAEC was detected in 37 (41.1%) cases involving diarrhoegenic E. coli. Studies conducted in Thailand and Brazil, reported a frequency of 12% and 11% EAEC respectively among children with acute diarrhoea [32, 33]. Although the prevalence of EAEC is believed to be considerably higher in the developing countries compared to industrially developed countries, a Switzerland study, reported that EAEC was encountered in a significant proportion of diarrhoea cases among children [34]. As evident in this study and previous studies, EAEC seems to be endemic within our study population and other locations in sub-Saharan Africa [35, 36], emerging as a significant diarrheal agent worldwide with the pattern of infection changing from persistent diarrhoea to include acute diarrhoea [37].
The second diarrheagenic E. coli type detected was EHEC constituting 21 (23.3%) of all diarrheagenic E. coli. Enterohaemorrhagic E. coli (EHEC) is a subset of Shiga toxin-producing Escherichia coli (STEC) which is associated with severe systemic disease as haemorrhagic colitis, haemolytic-uremic syndrome (HUS) and thrombotic thrombocytopenic purpura, particularly in infants, young children and in the elderly [38, 39]. EHEC infects the large bowel and inflict damage to the colon with infectious dose estimated to be less than 100 CFU [40]. Of the various virulence factors associated with pathogenicity in the EHEC strain, this study failed to detect both Shiga toxin 1 (stx1) and Shiga toxin 2 (stx2) but only detected intimin which is encoded by the eaeA gene [41]. Intimin is known to facilitate the adherence of pathogen to intestinal villi producing attaching and effacing lesions [42]. Previous studies have implicated EHEC in outbreaks and sporadic infections both in the United States and around the world [43, 44].
Intimin (eaeA) gene was also detected in 21 (23.3%) of EPEC. Unlike most studies where ETEC often take the lead in bacterial enteritis due to E. coli, a study by Weggerhof [45] also reported a higher incidence of EPEC from the screening of some pediatric patients with diarrhoea in Mpumalanga Province of South Africa. More recent studies described the contributions of EPEC to the human disease burden as significant [6, 46]. Thus EPEC plays a vital role in acute diarrhoea. EPEC is known to cause illness manifesting as watery diarrhoea with little inflammation of the intestinal mucosa [47]. Virulence is initiated in EPEC by the induction of a characteristic ultrastructural lesion in which the bacteria make intimate contact with the apical plasma membrane, causing localized destruction of the intestinal brush border and distortion of the apical enterocyte membrane [48] as is in the classical attaching and effacing (AE) lesion.
The ETEC and EIEC strains were found only in 9 (10.0%) and 2 (2.2%) of cases with diarrheagenic E. coli respectively. Although ETEC strains have been described as a major contributor to infantile diarrhoea in developing countries and of travellers' diarrhoea in visitors to these countries [49, 50], our findings were different showing a decline in the involvement of these strains in our setting. ETEC strains cause secretory diarrhoea similar to that of Vibrio cholerae by forming plasmid encoded heat-labile (LT) or heat-stable (ST) enterotoxins genes [47, 51]. ETEC engage strain-specific antigenic, hair-like fimbriae in attachment to specific receptors on the surface of enterocytes in the intestinal lumen [50]. EIEC on the other hand produce dysentery-like diarrhoea similar to that caused by Shigella species by invading and multiplying within epithelial cells of the colonic mucosa, resulting in an intense inflammatory response characterized by abscesses and ulcerations that damage the integrity of the epithelial cell lining of the colon [52]. EIEC was not a major enteric bacterial pathogen observed in this study, the prevalence was the least (2.2%) and this was similar to that obtained in the study conducted in Mexico City by Paniagu et al., [53] where EIEC was the least detected in the patient group (1%). This pattern is not consistent with studies in other developing countries where EIEC strains were important causes of pediatric diarrhea and dysentery [54, 55].
Salmonella species are an important cause of varying food and water-related infections. This study detected Salmonella as a major cause of gastroenteritis in our setting. Salmonella has previously been described as one of the common causes of gastroenteritis particularly in the developing countries [56, 57]. On the contrary, infectious diarrhoea in the developed world is often due to viruses [58]. The most common species isolated in this study were S. typhi (52%) and S. enterica serovar Typhimurium (25%). This report is consistent with other studies conducted in Iran and South Africa where S. typhi and S. enterica serovar Typhimurium were described as major aetiological agents of infectious diarrhoea [58, 59]. S. enterica serovar Isangi was third in ranking of frequency of isolation. Kruger et al., [60] described the increasing importance of this serotype of non-typhoidal Salmonella (NTS) which was a rare serotype in South Africa until 2002. Other species identified were S. enterica serovar Choleraesuis, S. enterica serovar Enteritidis, S. enterica serovar Eppendorf, S. enterica serovar Hadar, S. enterica serovar Panama and untyped Salmonella spp. The virulence factor detected among the majority (105) of the Salmonella spp. was inv A. This gene which is chromosomally located aids attachment of the pathogen to the epithelial cells [8]. The other detectable virulent gene was flic detected in 15 isolates. The flagellin gene, fliC is known to aid systemic spread of pathogen and is specific for S. enterica serovar Typhimurium [61]. Enteric bacteria possessing sefA which encodes the SEF14 fimbrial antigen, a virulence plasmid specific for S. enterica serovar Enteritidis [62] were not encountered jn this study.
Conclusions
This study showed the diversity of virulence gene expression in two major enteric pathogens. It was observed among other things that some diarrhoegenic E. coli isolated from apparently asymptomatic subjects expressed some virulence genes at frequency as high as seen in diarrhoegenic cases. This is a pointer to the fact that asymptomatic individuals serve as reservoirs of pathogenic strains of enteric bacteria and may play a role in the spread and acquisition of virulence genes.
References
Amar CF, East CL, Gray J, Iturriza-Gomara M, Maclure EA, McLauchlin J: Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993-1996). Eur J Clin Microbiol Infect Dis. 2007, 26: 311-323. 10.1007/s10096-007-0290-8.
Gordon MA: Salmonella infections in immunocompromised adults. J Infect. 2008, 56: 413-422. 10.1016/j.jinf.2008.03.012.
Falkow S: The evolution of pathogenicity in Escherichia, Shigella, and Salmonella. Escherichia coli and Salmonella: cellular and molecular biology..2 edition. Edited by: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB Jr, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. American Society for Microbiology (ASM) Press, Washington, D.C; 1996:2723-2729.
Selander RK, Musser JM: Population genetics of bacterial pathogenesis. Molecular Basis of Bacterial Pathogenesis. Edited by: Iglewski BH, Clark VL. 1990, Academic Press, Inc., San Diego, Calif, 11-36.
Finlay BB, Falkow S: Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997, 61: 136-169.
Kaper JB, Nataro JP, Mobley HL: Pathogenic Escherichia coli. Nat Rev Microbiol. 2004, 2: 123-140. 10.1038/nrmicro818.
Altier C: Genetic and Environmental Control of Salmonella Invasion. J Microbiol 2005, 43: 85-92, special issue.
Galán JE, Curtiss R: Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989, 86: 6383-6387. 10.1073/pnas.86.16.6383.
McCormick BA: Invasion mechanisms of Salmonella. Advances in Molecular and Cellular Microbiology. Bacterial Invasion of Host Cells. Edited by: Richard J Lamont. Cambridge University Press. The Edinburgh Building, Cambridge CB2 2RU, UK; 2004:1-24.
Buchmeier NA, Heffron F: Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect Immun. 1991, 59: 2232-2238.
Pizarro-Cerdà J, Moreno E, Desjardins M, Gorvel JP: When intracellular pathogens invade the frontiers of cell biology and immunology. Histol Histopathol. 1997, 12: 1027-1038.
Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P: Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001, 2: 361-367. 10.1038/86373.
Vasquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parkos WT, Fang FC: Extraintestinal dissemination of Salmonella by CD-18 expressing phagocytes. Nature. 1999, 401: 804-808. 10.1038/44593.
Hacker J, Hentschel U, Dobrindt U: Prokaryotic Chromosomes and Disease. Science. 2003, 301: 790-793. 10.1126/science.1086802.
Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J: Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wildtype pathogen. Infect Immun 1994, 62: 606-614.
Mahan MJ, Slauch JM, Mekalanos JJ: Environmental regulation of virulence gene expression in Escherichia, Salmonella and Shigella spp. Escherichia coli and Salmonella: cellular and molecular biology..2 edition. Edited by: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB Jr, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. American Society for Microbiology (ASM) Press, Washington, D.C.; 1996:2803-2815.
Mellata M, Ameiss K, Mo H, Curtiss R: Characterization of the Contribution to Virulence of Three Large Plasmids of Avian Pathogenic Escherichia coli χ7122 (O78:K80:H9). Infect Immun. 2010, 78 (4): 1528-1541. 10.1128/IAI.00981-09.
Galan JE, Wolf-Watz H: Protein delivery into eukaryotic cells by type III secretion machines. Nature 2006, 444: 567-573. 10.1038/nature05272.
Collazo CM, Zierler MK, Galan JE: Functional analysis of the Salmonella typhimurium invasion genes invL and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol Microbiol. 1995, 15: 25-38. 10.1111/j.1365-2958.1995.tb02218.x.
Naum M, Brown EW, Mason-Gamer RJ: Phylogenetic evidence for extensive horizontal gene transfer of type III secretion system genes among enterobacterial plant pathogens. Microbiology 2009, 155: 3187-3199. 10.1099/mic.0.029892-0.
Guo A, Lasaro MA, Sirard J, Kraehenbu J, Schifferli DM: Adhesin-dependent binding and uptake of Salmonella enterica serovar Typhimurium by dendritic cells. Microbiology 2007, 153: 1059-1069. 10.1099/mic.0.2006/000331-0.
Musser JM: Molecular Population Genetic Analysis of Emerged Bacterial Pathogens: Selected Insights. Emerging Infect Dis 1996, 2: 1-17. 10.3201/eid0201.960101.
Bansal AK: Role of bioinformatics in the development of new antibacterial therapy. Expert Rev Anti-infective Therap 2008, 6 (1): 51-65. 10.1586/14787210.6.1.51.
Gonzalez-Escobedo G, Marshall JM, Gunn JS: Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nature Rev Microbiol 2011, 9: 9-14. 10.1038/nrmicro2490.
Forbes BA, Sahm DF, Weissfeld A: Diagnostic Microbiology. Bailey & Scott's Diagnostic Microbiology. 11 edition. Mosby Publisher; 2002, 1069.
Mandeville KL, Krabshuis J, Ladep NG, Mulder CJJ, Quigley EMM, Khan SA: Gastroenterology in developing countries: Issues and advances. World J Gastroenterol 2009, 15 (23): 2839-2854. 10.3748/wjg.15.2839.
Bradshaw D, Nannan N, Laubscher R, Groenewald P, Joubert J, Nojilana B, Norman R, Pieterse D, Schneider M: South African National Burden of Disease Study, 2000: Estimates of Provincial Mortality. Tygerberg, SAMRC; 2004.
Rispel L, Setswe G: Stewardship: Protecting the public's health. South African Health Review. Edited by: Harrison S, Bhana R, Ntuli A. Durban: Health Systems Trust; 2007:, URL: http://www.hst.org.za/publications/711.
Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Cieslak PR, Deneen VC, Tauxe RV: FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis 2004, 38 (3): S127-S134. 10.1086/381578.
Galan JE, Sansonetti PJ: Molecular and cellular bases of Salmonella and Shigella interactions with host cells. Escherichia coli and Salmonella: cellular and molecular biology..2 edition. Edited by: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB Jr, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. American Society for Microbiology (ASM) Press, Washington, D.C.; 1996:2757-2773.
Tang C, Holden D: Pathogen virulence genes - implications for vaccines and drug therapy. British Med Bull 1999, 55 (2): 387-400. 10.1258/0007142991902448.
Souza EC, Martinez MB, Taddei CR, Mukai L, Gilio AE, Racz ML, Silva L, Ejzenberg B, Okay Y: Perfil etiologico das diarreias agudas de criancas atendidas em Sao Paulo. Etiologic profile of acute diarrhea in children in Sao Paulo. J de Pediatria 2002, 78 (1): 31-38.
Ratchtrachenchai OA, Subpasu S, Hayashi H, Ba-Thein W: Prevalence of childhood diarrhoeaassociated Escherichia coli in Thailand. J Med Microbiol 2004, 53 (3): 237-243. 10.1099/jmm.0.05413-0.
Pabst WL, Altwegg M, Kind C, Mirjanic S, Hardegger D, Nadal D: Prevalence of enteroaggregative Escherichia coli among children with and without diarrhea in Switzerland. J Clin Microbiol 2003, 41: 2289-2293. 10.1128/JCM.41.6.2289-2293.2003.
Geyid A, Olsvik O, Ljungh A: Virulence properties of Escherichia coli isolated from Ethiopian patients with acute or persistent diarrhoea. Ethiop Med J 1998, 36: 123-139.
Okeke IN, Ojo O, Lamikanra A, Kaper JB: Etiology of Acute Diarrhea in Adults in Southwestern Nigeria. J Clin Microbiol 2003, 41 (10): 4525-4530. 10.1128/JCM.41.10.4525-4530.2003.
Scaletsky ICA, Fabbricotti SH, Silva SOG, Morais MB, Fagundes-Nieto U: HEp-2-adherence Escherichia coli strains associated with acute infantile diarrhea, Sao Paulo, Brazil. Emerg Infect Dis 2002, 8: 855-858.
Nataro JP, Kaper JB: Diarrheagenic Escherichia coli. Clin Microbiol Rev 1998, 11: 142-201.
Paton AW, Paton JC: Detection and characterization of Shiga Toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfb O111, and rfb O157. J Clin Microbiol 1998, 36: 598-602.
Mellies JL, Barron AMS, Anna M: Enteropathogenic and Enterohemorrhagic Escherichia coli Virulence Gene Regulation. Infect Immun 2007, 9 (75): 4199-4210.
Kang SJ, Ryu SJ, Chae JS, Eo SK, Woo GJ, Lee JH: Occurrence and characteristics of enterohemorrhagic Escherichia coli O157 in calves associated with diarrhoea. Vet Microbiol 2004, 98: 323-328. 10.1016/j.vetmic.2003.11.004.
Caprioli A, Morabito S, Brugere H, Oswald E: Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet Research 2005, 36: 289-311. 10.1051/vetres:2005002.
Waters JR, Sharp JC, Dev VJ: Infection caused by Escherichia coli O157:H7 in Alberta, Canada, and in Scotland: a five-year review, 1987-1991. Clin Infect Dis 1994, 19: 834-10.1093/clinids/19.5.834.
Dundas S, Todd WT, Stewart AI, Murdoch PS, Chaudhuri AKR, Hutchinson SJ: The central Scotland Escherichia coli O157:H7 outbreak: risk factors for the hemolytic uremic syndrome and death among hospitalized patients. Clin Infect Dis 2001, 33: 923-10.1086/322598.
Weggerhof FO: Masters dissertation. The aetiology of gastroenteritis in infants in a rural population. University of the Witwatersrand, Johannesburg, South Africa; 1987.
Scaletsky SCA, Souza TB, Aranda KRS, Okeke IN: Genetic elements associated with antimicrobial resistance in enteropathogenic Escherichia coli (EPEC) from Brazil. BMC Microbiol. 2010, 10: 25-10.1186/1471-2180-10-25.
Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC, Eds: Color atlas and textbook of diagnostic microbiology. Lippincott: Philadelphia, New York, United States of America;, Fifth 1997, 171-199.
Clarke SC, Haigh RD, Freestone PPE, Williams PH: Virulence of Enteropathogenic Escherichia coli, a Global Pathogen. Clin Microbiol Rev 2003, 16 (3): 365-378. 10.1128/CMR.16.3.365-378.2003.
Sooka A, du Plessis M, Keddy K: Enterovirulent Escherichia coli. Southern Afr J Epidemiol Infect 2004, 19 (1): 23-33.
WHO World Health Organization: Diarrhoeal Diseases. Enterotoxigenic Escherichia coli (ETEC) 2009 [http://www.who.int/vaccine_research/diseases/diarrhoeal/en/index4.html], (Updated February 2009).
Levine MM: Enteric infections and the vaccines to counter them: future directions. Vaccine 2006, 24: 3865-3873. 10.1016/j.vaccine.2006.03.039.
Hale TL: Bacillary dysentery.In Topley and Wilson's microbiology and microbial infections. Volume 3. Edited by: Hansler WJ, Shuman M. Arnold, London, England; 1998:479-493.
Paniagua GL, Monroy E, García-González O, Alonso J, Negrete E, Vaca S: Two or more enteropathogens are associated with diarrhoea in Mexican children. Annals Clin Microbiol Antimicrob 2007, 6: 17-10.1186/1476-0711-6-17.
Taylor DN, Echeverria P, Pal T, Sethabutr O, Saiborisuth S, Sricharmorn S, Rowe B, Cross J: The role of Shigella spp., enteroinvasive Escherichia coli, and other enteropathogens as causes of childhood dysentery in Thailand. J Infect Dis 1986, 153: 1132-1138. 10.1093/infdis/153.6.1132.
Pacheco-Gil L, Ochoa TJ, Flores-Romo L, DuPont HL, Estrada-Garcia T: Enteroinvasive Escherichia coli severe dysentery complicated by rotavirus gastroenteritis. J Infect 2006, 53 (5): e211-e213. 10.1016/j.jinf.2006.01.021.
Guerrant R, Hughes J, Lima N, Crane J: Diarrhea in developed and developing countries: magnitude, special settings, and etiologies. Rev Infect Dis 1990, 1: 41-50.
Galanis E, Lo Fo Wong DM, Patrick ME, Binsztein N, Cieslik A, Chalermchikit T, Aidara-Kane A, Ellis A, Angulo FJ, Wegener HC: World Health Organization Global Salm-Surv. Web-based surveillance and global Salmonella distribution 2000-2002. Emerg Infect Dis 2006, 12: 381-388.
Jafari F, Shokrzadeh L, Hamidian M, Salmanzadeh-Ahrabi S, Zali MR: Acute diarrhoea due to enteropathogenic bacteria in patients at hospitals in Tehran. Jpn J infect Dis 2008, 61: 269-273.
Keddy KH, Dwarika S, Crowther P, Perovic O, Wadula J, Hoosen A, Sooka A, Crewe-Brown HH, Smith AM: Genotypic and demographic characterization of invasive isolates of Salmonella Typhimurium in HIV co-infected patients in South Africa. J Infect Dev Ctries 2009, 3 (8): 585-592.
Kruger T, Szabo D, Keddy KH, Deeley K, Marsh JW, Hujer AM, Bonomo RA, Paterson DL: Infections with Nontyphoidal Salmonella Species Producing TEM-63 or a Novel TEM Enzyme, TEM-131, in South Africa. Antimicrob Agents Chemother 2004, 48 (11): 4263-4270. 10.1128/AAC.48.11.4263-4270.2004.
Soumet C, Ermel G, Rose V, Rose N, Drouin P, Salvat G, Colin P: Identification by a multiplex PCRbased assay of Salmonella Typhimurium and Salmonella Enteritidis strains from environmental swabs of poultry houses. Let Applied Microbiol 1999, 29: 1-6. 10.1046/j.1365-2672.1999.00559.x.
Doran JL, Collinson SK, Clouthier SC, Cebula TA, Koch WH, Burian J, Banser PA, Todd ECD, Kay WW: Diagnostic potential of sefA DNA probes to Salmonella Enteritidis and certain other O-serogroup D1 Salmonella serovars. Mol Cellular Probes 1996, 10: 233-246. 10.1006/mcpr.1996.0033.
Wang G, Clark CG, Rodgers FG: Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol 2002, 40: 3613-3619. 10.1128/JCM.40.10.3613-3619.2002.
Jothikumard N, Griffiths MW: Rapid detection of Escherichia coli O157:H7 with multiplex real-time PCR assays. Appl Environ Microbiol 2002, 68: 3169-3171. 10.1128/AEM.68.6.3169-3171.2002.
Ito F, Hagino T, Ito KI, Watanabe H: Differentiation and detection of pathogenic determinants among diarrheagenic Escherichia coli by polymerase chain reaction using mixed primers. Nihonrinshou 1992, 50: 343-347, (In Japanese).
Franck SM, Bosworth BT, Moon HW: Multiplex PCR for enterotoxigenic, attaching and effacing, and Shiga toxin-producing Escherichia coli strains from calves. J Clin Microbiol 1998, 36: 1795-1797.
Ratchtrachenchai OA, Subpasu S, Ito K: Investigation on enteroaggregative Escherichia coli infection by multiplex PCR. Bull Dept Med Sci 1997, 39: 211-220.
Yatsuyanagi J, Saito S, Sato H, Miyajima Y, Amano KI, Enomoto K: Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J Clin Microbiol 2002, 40 (1): 294-297. 10.1128/JCM.40.1.294-297.2002.
Villalobo E, Torres A: PCR for detection of Shigella spp. in mayonnaise. Appl Environ Microbiol 1998, 64: 1242-1245.
Rahn K, De Grandis SA, Clarks RC, McEwen SA, Galan JE, Ginocchio C, Curtis R, Gyles CI: Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes 1992, 6: 271-279. 10.1016/0890-8508(92)90002-F.
Soumet C, Ermel G, Rose V, Rose N, Drouin P, Salvat G, Colin P: Simultaneous detection by PCR of Salmonella Typhimurium and Salmonella Enteritidis from environmental samples of poultry houses. Salmonella and Salmonellosis Proceedings. Edited by: Colin P, Le Goux JM, Clement G. Ploufragan, France: ISPAIA; 1997:53-57.
Joys TM: The covalent structure of the phas-1 flagellar filament protein of Salmonella Typhimurium and its comparison to others flagellins. J Biol Chem 1985, 260: 15758-15761.
Acknowledgements
This study was supported by the Institutional Research Grant of Walter Sisulu University (WSU), the National Research Foundation (NRF) grant awarded under the aegis of the South Africa - Japan Research Collaborative Agreement and the Focus Area Grant of the NRF. Our profound gratitude goes to Dr. Karen Keddy, Arvinda Sooka and other members of Enteric Diseases Reference Unit of the National Institute of Communicable Diseases, Johannesburg for their support during isolate collection. We also appreciate the technical support of Christiaan Labuschagne (Inqaba Biotechnology, Pretoria), Dr Li (Department of Emerging Infectious Diseases Unit, Tohoku University, Sendai, Japan) and Benjamin (University of Venda, South Africa) for molecular analysis and interpretation. The general support obtained from the Department of Medical Microbiology, National Health Laboratory Services, Nelson Mandela Academic Hospital, Mthatha is worthy of commendation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MAB participated in the design of the study, carried out laboratory analysis and drafted the manuscript. CLO conceived of the study, participated in the design and coordination of the study, supervised the study and revised the manuscript. TH coordinated bench work between collaborators in South Africa and Japan and helped to revise the manuscript. KAB was involved in coordination and facilitated activities at NICD. SDV assisted with the concept and design of the study. Authors read and approved the final manuscript.
Electronic supplementary material
13099_2011_45_MOESM1_ESM.PPT
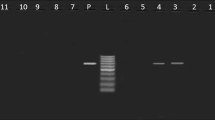
Additional file 1:Representative gels for PCR amplification of DNA extracted from selected E. coli isolates showing the presence of diverse virulence genes. A: 100 bp molecular weight marker (lanes 1 and 10), fragment from agg R (lanes 2 to 4), eaeA (lanes 5 to 6) and astA (lanes 7 to 9). B: 100 bp molecular weight marker (lanes 1 and 6), fragment from agg R (lanes 2 to 3), virA (lanes 4) and LT (lanes 5). The relative positions in the gel of predicted size of PCR products are indicated by arrowheads on the right sides. (PPT 180 KB)
13099_2011_45_MOESM2_ESM.PPT
Additional file 2:Representative gels for PCR amplification of DNA extracted from selected Salmonella isolates showing the presence of diverse virulence genes. 100 bp molecular weight marker (lanes 1 and 8) fli C (lanes 2 to 4) and inv A (lanes 5 to 7). The relative positions in the gel of predicted size of PCR products are indicated by arrowheads on the right sides. The relative positions in the gel of predicted size of PCR products are indicated by arrowheads on the right sides. (PPT 106 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bisi-Johnson, M.A., Obi, C.L., Vasaikar, S.D. et al. Molecular basis of virulence in clinical isolates of Escherichia coli and Salmonella species from a tertiary hospital in the Eastern Cape, South Africa. Gut Pathog 3, 9 (2011). https://doi.org/10.1186/1757-4749-3-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1757-4749-3-9