Abstract
Flaviviruses are enveloped single positive-stranded RNA viruses. The capsid (C), a structural protein of flavivirus, is dimeric and alpha-helical, with several special structural and functional features. The functions of the C protein go far beyond a structural role in virions. It is not only responsible for encapsidation to protect the viral RNA but also able to interact with various host proteins to promote virus proliferation. Therefore, the C protein plays an important role in infected host cells and the viral life cycle. Flaviviruses have been shown to affect the health of humans and animals. Thus, there is an urgent need to effectively control flavivirus infections. The structure of the flavivirus virion has been determined, but there is relatively little information about the function of the C protein. Hence, a greater understanding of the role of the C protein in viral infections will help to discover novel antiviral strategies and provide a promising starting point for the further development of flavivirus vaccines or therapeutics.
Similar content being viewed by others
1 Introduction
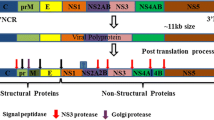
Flavivirus is part of the Flaviviridae family along with Pestivirus, Hepacivirus and Pegivirus [1, 2]. The primary categories are mosquito-borne (such as Usutu virus, USUV; dengue virus, DENV; Japanese encephalitis virus, JEV; West Nile virus, WNV; Zika virus, ZIKV) and tick-borne flaviviruses (such as tick-borne encephalitis virus, TBEV). These flaviviruses mainly cause foetal malformation (such as ZIKV) and neurological disorders (such as JEV and TBEV). A flavivirus is an enveloped single positive-strand RNA virus with a genome of approximately 11 kb that contains a single open reading frame (ORF) flanked with a short (~100 nt) 5′-untranslated region (UTR) and a longer (~400–600 nt) 3′-UTR [3]. The ORF encodes a polyprotein associated with the endoplasmic reticulum (ER) membrane and is proteolytically cleaved into three structural proteins (capsid, C; precursor membrane, prM; and envelope, E) and at least seven nonstructural (NS) proteins (NS1, NS2A/B, NS3, NS4A/B and NS5) by host and viral proteases [4]. The C protein is essential for the assembly and maturation of viral particles and is the most promising drug target candidate. Before being cleaved by a viral protease to produce a mature protein, the C protein takes the form of a membrane-anchored C (anchC), which can initiate subsequent effective flavivirus assembly but will not perform viral replication [5], and this process induces the downstream prM to be cleaved into Pr and M by furin [6]. anchC is also the signal peptide that transfers prM to the ER lumen. Therefore, anchC is produced and disappears in the life cycle of flavivirus, and its mechanism of action in virus assembly may be helpful for antiviral research. However, the size and transient existence of anchC may limit its research. The nonstructural proteins are proteases that are mainly involved in the cleavage of polyprotein [7] and the regulation of host cell responses [8]. NS2A and NS3 also participate in the assembly of virions through direct interaction with the C protein [9]. NS1 regulates the production of infectious particles by interacting with structural proteins (C, prM and E) [10, 11].
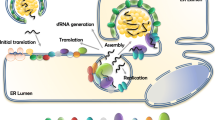
Flaviviruses use a complex reproduction process to gain access to host cells (Figure 1). Flavivirus infection can induce invagination of the ER, forming clusters of double-membrane vesicles (Ves) wrapped in vesicle packets (VPs) [12]. The Ve houses the viral replication complex, including double-stranded viral RNA, viral nonstructural proteins and cellular proteins [13]. C protein binds to the membrane and exits the ER via a coat protein complex II (COPII)-dependent mechanism, bypassing the Golgi apparatus; the presence of interferon-induced protein viperin enhances the release of C protein [14]. Currently, there is relatively little information on the functional properties of flavivirus C protein. However, C protein is the critical element of infectious virus particles and contains positively charged residues distributed throughout the molecule. The mature C protein assembles on the genomic RNA through nonspecific electrostatic interactions to form a nucleocapsid, which is wrapped by the lipid bilayer with prM and E proteins to form an infectious virion [15, 16]. The purified C protein dimer can be assembled into C-like particles when combined with transcribed viral RNA in vitro [17], and the C protein-coding region hairpin sequence (cHP) is a conserved region in arthropod-borne flaviviruses. cHP is involved in the enhanced recognition of translation initiation codons, which are essential for effective RNA replication [18]. In this review, we primarily summarize the structure and function of the C protein, aiming to discover more new functions and assist in the development of flavivirus vaccines and drugs.
Propagation process of flaviviruses. Flavivirus propagation mainly includes adsorption, entrance, replication, assembly, maturity and release. The virus enters host cells through host receptor recognition (A); then, the virus fuses with the host membrane (B), and the RNA of flavivirus is ejected from the nucleocapsid and released into the cytoplasm of the host cells (C). RNA replication, protein synthesis and particle assembly are carried out in the ER (D and E); the particles mature in the Golgi apparatus (F); subsequently, mature virions are released from the host cells (G).
2 Structure of the C protein
The structure of the C protein has been solved by nuclear magnetic resonance (NMR) and X-ray crystallography [19]. The C protein is located at the N-terminus of the polyprotein [20]; it is an internal protein in virus structure, and we expect it to consist of approximately 180 capsid protein units [21]. The C protein is composed of approximately 100 amino acid residues and has a molecular weight of approximately 13 kDa; for example, the C protein of Tembusu virus (TMUV) consists of 120 amino acids (aa), that of WNV contains 105 aa, and that of DENV contains 100 aa. Compared with the other two surface proteins prM and E in different flaviviruses, the C protein has the lowest amino acid homology. The homology of the JEV C protein to WNV, DENV-2 and TBEV is only 67%, 33%, and 25%, respectively. However, certain characteristics, such as the biochemical properties, structural specifics (hydrophobicity, secondary/tertiary structure, and abundance of basic amino acid residues) [22] and several functional elements of the C protein, seem to be well conserved in flaviviruses [23, 24]. These conservative biochemical characteristics contribute to similar functions.
2.1 Superhelical structure of the C protein
Each C protein molecule contains two highly conserved internal regions: a hydrophobic region and a highly cationic region [25]. A far-ultraviolet circular dichroism (CD) potassium analysis showed that the flavivirus C protein monomer forms oligomers in solution and that the protein is predominantly a dimer with a two–alpha helical conformation, including four α-helices (α1 to α4) connected by short loops [26] (Figure 2). The C protein dimer is essential for viral assembly and virus particle stability [27]. The α4 helix at the hydrophobic C-terminus of the C protein is the longest of the four helices, and the α4–α4′ interaction plays the most important role in supporting C protein dimers [28], nucleocapsid formation and virus production [29]. The α1–α1′ and α2–α2′ helices are located on the opposite sides of the α4-α4′ helix and are composed of nonpolar residues [30]. Some researchers think that the α4–α4′ region interacts with RNA, and the hydrophobic core in the α2–α2′ region binds to the virus and host lipid membranes [31]. The α1 helix is a part of the central hydrophobic region, which may play an important role in virus assembly and interactions with virus surface proteins [32, 33]. The α3–α3′ helix is parallel to the α4–α4′ helix, and the α1 and α3 helices are amphipathic and are mainly composed of leucine residues.
3D structure of the flavivirus capsid protein. Topological diagram of a C protein homodimer: α-helices 1–4 of each C monomer are shown in light blue, green, purple and dark blue, respectively. The α1–α1′ and α2–α2′ helices are located on the opposite side of the α4–α4′ helices, and the α3–α3′ helix is parallel to the α4–α4′ helix. The 3D structure was created with PyMOL software.
2.2 Nucleocapsid
The flavivirus particle consists of an electron-dense inner core called the nucleocapsid (NC). The NC complex contains multiple copies of the C protein and a single copy of genomic RNA, and the production of infectious virus particles requires the NC. The anchC may help initiate the formation of the nucleocapsid, including interactions with the genomic RNA. The mature C protein is responsible for packaging the viral nucleic acid and ribonucleoprotein complex in the virion, and RNA encapsidation is the first step in the assembly process of flaviviruses [34]. In the process of encapsidation, the C protein acts as a chaperone of RNA, promoting the folding of RNA by preventing its misfolding or by dissolving misfolded RNA without consuming ATP. The C protein is also involved in regulating the cyclization of flavivirus genome RNA for viral replication [35]. The NC is enveloped by two other structural proteins (prM/E) and the lipid bilayer derived from the host cell ER [36,37,38] (Figure 3). The interaction between the C protein and nucleolin in the nucleolus is important for the formation of stable and functional nucleocapsids [39]. During the virus infection cycle, the dissociation mode of the NC in the cytoplasm is not yet completely understood and may be worth exploring in antiviral studies.
2.3 Nuclear localization signals
The C protein of flavivirus is localized in both the cytoplasm and the nucleus. The C protein has basic amino acid regions (mainly 23–25% Arg and Lys) called nuclear localization signals (NLSs), and it can interact with nuclear import receptor proteins (such as transcription factor hnRNP K, nucleophosmin and importin α/β), which promote entry of the C protein into the nucleus; the NLSs of WNV are located from aa 85 to aa 101 [40], and the NLSs of DENV are located from aa 85–100, including 6KKAR9, 73KKSK76, and the bipartite signal 85RKeigrmlnilnRRRR100 [41,42,43]. The active transport system may be related to the movement of C protein from the cytoplasm to the nucleus, and importins (α/β), protein kinase and C protein-mediated phosphorylation are involved in this process [40]. The interaction between the C protein and importin-α is mediated by NLS motifs, but the function of the C protein in the nucleolus is almost unknown. According to evolutionary study of the Flaviviridae, C protein NLSs are highly conserved in mosquito-borne flavivirus and blood-borne and human-adapted hepatitis C virus (HCV). The Gly42 and Pro43 of C proteins are important for nuclear localization and are completely conserved in flavivirus and HCV [44]. After mutating Gly42 and Pro43 to Ala (the M4243 mutant), an increase in defective particles or a low viral RNA replication efficiency was detected in JEV [45].
2.4 Phosphorylation sites
The NetPhos algorithm shows that flavivirus C protein has 2–6 putative phosphorylation sites. These phosphorylation sites exist in the RNA-binding regions of the C protein. Therefore, dephosphorylation of the C protein enhances its interaction with viral RNA. According to bioinformatics analyses, the WNV C protein is phosphorylated in infected cells and has 5 putative phosphorylation sites (serine 26, 36, 83, 99 and threonine 100). The work of Cheong [46] showed that mutation in these phosphorylation sites of the C protein reduced its RNA binding activity and did not inhibit oligomerization but did affect the ratio of dimerization and oligomerization. In infected cells, the flavivirus C protein is localized in the nucleus due to the presence of NLSs. Interestingly, the degree of phosphorylation of the C protein is reduced, thereby reducing its nuclear localization [47].
2.5 Structural flexibility of the C protein
The N-terminal region of the C protein is unstructured in solution, especially the first 20 residues [48]. This region has abundant positive charges because it is rich in the basic residues Arg and Lys. Therefore, almost 40% of the unstructured N-terminus of the C protein can be removed without severely damaging its functional integrity. Hence, truncation of the N-terminal region has been used in the prokaryotic expression and purification of recombinant C. The truncated C protein retains the ability to package RNA, and even the version with approximately 30% of the C-terminal region deleted (including complete deletion of α4) can still function [27]. Removing the entire internal hydrophobic domain from the α2 helix and the partial loop between the α2 and α3 helices of the WNV C protein severely impairs viral growth; however, a short deletion did not substantially affect the growth, indicating the structural and functional flexibility of the C protein [44, 49]. A TBEV study showed that the C-terminal region of the C protein has two amino acid sequence motifs that match the canonical NS2B/NS3 recognition site, and this region has significant functional flexibility in the assembly of infectious virions [50]. The flexibility of the C protein is conferred by its inherent disordered region and is conserved in all flavivirus C proteins. The key to the many actions that C proteins can perform in the proliferation cycle of virus particles and cells is the existence of internal disordered domains.
2.6 Cis-acting elements
Cis-acting elements are necessary for genome circularization and viral enzyme activity in replication, and the rate of viral replication is very sensitive to small changes in this RNA. The N-terminus of the C protein-coding region is important in the translation, replication, host adaptation and encapsidation of mosquito-borne flavivirus [29, 51]. A novel cis-acting element downstream of the 5′ cyclization sequence pseudoknot (DCS-PK) is conserved in mosquito-borne flaviviruses [52], which mainly enhances flavivirus RNA replication by regulating genome cyclization. The function of DCS-PK depends mainly on its secondary structure and some conserved primary sequences, such as the highly conserved stem 1 loop 2 sequence [53]. Stem-loop 6 (SL6) is a cis-acting enhancer in the C protein coding region of tick-borne flavivirus [54]. Cis-acting elements can be used to study the function of flavivirus C proteins.
3 Multiple functions of the C protein
The multiple functions of the flavivirus C protein are attributed to the abovementioned special structural features (Figure 4). The C protein undergoes various conformational changes and participates in the formation of the nucleocapsid to protect viral RNA. It also has nonstructural functions in the virus life cycle, mainly involving intermolecular interactions, such as interaction with organelle membranes to promote virus replication, assembly and virion maturation.
3.1 Binding and interacting with RNA
The NS2A protein of flavivirus can recruit genomic RNA, structural proteins (C/prM/E), and nonstructural proteins (NS2B/NS3) to the virion assembly site. Once the C-prM-E polyprotein is formed, NS2A will transfer the viral RNA to the structural proteins for virion assembly [36]. The C protein binds and interacts with RNA to form a nucleocapsid and is wrapped by prM/E proteins. The mutation of NS2A weakens the binding and eliminates virus production, which indicates that NS2A plays an indispensable role in virion assembly [55].
Cell-penetrating peptides (CPPs) can translocate to the cell membrane and function to reduce immunogenicity and cytotoxicity. Recent publications indicate that CPPs can deliver many bioactive molecules such as proteins, nucleic acids, and therapeutics in mammalian cells [56]. CPPs derived from the DENV C protein are involved in suitable viral delivery systems, which can deliver nucleic acids to cells for virus infection or deliver therapeutic molecules [21, 57]. CPPs contain 6–30 amino acid residues, which range from highly charged (rich in Arg and Lys) to hydrophobic or amphipathic sequences. Basic residues are cumulatively distributed on the surface of the C monomer; the positive charges of these basic residues may play a role in the neutralization of negative charges during viral RNA encapsidation. To assemble the nucleocapsid and package the genome without nucleases, the hydrophobic amino acids at the carboxyl terminus of the C protein are fixed to the rough ER of the host cell.
The structural part of the DENV C protein includes two domains, pepR and pepM, which have very distinct physical and chemical characteristics [21]. They are two novel intrinsic CPPs that have conserved folds in the C proteins of the Flaviviridae family [58], and they have two different internalization routes. pepR uses the endocytic pathway, while pepM directly translocates physically through the lipid membrane. These two peptides preferentially bind to anionic lipid membranes in an α-helical configuration [21]. pepM is a relatively hydrophobic domain, while pepR is a highly cationic helix. Therefore, pepM primarily interacts with lipid membranes, and pepR binds to RNA in the virion. However, only pepM can promote the fusion and aggregation of Ve, which is composed of zwitter-ionic lipids [59]. The C protein of yellow fever virus (YFV) has a high-affinity dsRNA-binding function, which can interfere with the cleavage of long dsRNA by Dicer, thereby antagonizing RNA silencing [60]. The conservation of the viral suppressor function in various flavivirus C proteins is particularly fascinating. The basic residues in the N-terminal region help RNA bind to form particles [61]. Viral protein-derived CPPs can be a valuable tool for drug delivery across membranes, mainly in genetic therapy. Therefore, researchers designed and synthesized pepM and pepR based on the two domains of the C protein sequence. They have almost equivalent efficiency to the whole parental C protein for ssDNA delivery. In addition to the CPP properties, pepR also has antibacterial activity.
Based on the combination of C protein and RNA, researchers have considered fusing exogenous genes with C protein through a capsid-targeted antiviral inactivation (CTVI) strategy [62]. This tactic is based on the fusion of the C protein and a crucial effector molecule, such as a nuclease, a lipase, a protease, or a single-chain antibody (scAb), which can degrade viral DNA/RNA or interfere with the proper folding of important viral proteins [63].
3.2 Promoting the proper assembly of infectious particles
The C-terminus of the C protein contains a single transmembrane domain (TMD) called anchC, which acts as a signal for the translocation of prM into the ER lumen and controls the stability of the E protein, so it is available for the assembly of infectious particles [64, 65]. The TMD is cleaved by the viral protease NS2B/NS3 to release a functional cytosolic form. In addition, due to the internal hydrophobic sequence (IHS), mature C protein can bind to cellular and intracellular membranes. IHS contains 14 to 22 hydrophobic residues, which are conserved in all flavivirus C proteins and are required for virus maturation and assembly [27]. The C and prM proteins are connected by the IHS, which crosses the ER membrane and is responsible for the translocation of prM to the ER lumen [50, 66]. In a TBEV study, Kofler interfered with the assembly of viral particles by deleting HIS [32], and the proportion of subviral particles (SVPs, lacking the nucleocapsid and infectivity) produced by deletion mutants increased. However, SVPs are highly immunogenic because of the prM and E proteins [67]. Moreover, the C protein plays a vital role in other aspects of the flavivirus replication cycle. Kofler et al. [32] proved that large deletions in the C protein (C △28–35, C △28–39, C △28–43, and C △28–48) did not significantly reduce the level of the C protein in BHK-21, and C protein deletion mutants did not impair RNA replication or translation. However, these mutations can affect the expression of the E protein, which indirectly affects the formation of particles. For example, the diameter of capsid-free and noninfectious SVPs is smaller than that of wild-type viral particles, the viral infectious titres of the four mutants are lower than that of wild-type virus, and the production of infectious particles released by all mutants is lower than that of wild-type virus.
3.3 Phosphorylation and dephosphorylation of the C protein affect virus replication
The phosphorylation of the C protein is an essential process of virus replication in many viruses. It is an important posttranscriptional modification required for flavivirus C protein function, such as binding to importin-α or HDM2 protein [47]. Experiments with kinase inhibitors and activators have shown that protein kinase C is responsible for the phosphorylation of the C protein [47]. C protein undergoes spontaneous self-oligomerization, which is involved in nucleocapsid formation; however, phosphorylation reduces both the oligomerization rate and phosphorylation-dependent nucleocapsid assembly [46]. Dephosphorylation of the C protein is also critical for reducing its nuclear localization. Therefore, during viral infection, the phosphorylation of the C protein should decrease over time. The assembly of flavivirus occurs in the cytoplasm. In the later stages of infection, dephosphorylation promotes the interaction of C protein with RNA in the cytoplasm. Tracking the cellular location of the C protein during infection indicated that it was localized in the nucleus in the early stages of infection and in the cytoplasm in the late phase of infection. These changes are related to the gradual dephosphorylation of C protein in infected cells. However, in Flaviviridae, only the C proteins of HCV and WNV have been shown to be phosphorylated. In fact, the bioinformatics analysis of other flavivirus C proteins revealed multiple putative phosphorylation sites. Therefore, the C proteins of other flaviviruses may also be phosphorylated.
3.4 Interactions with host proteins to promote virus propagation
The C protein binds to viral RNA and has other cellular regulatory functions in infected cells. This protein can mediate host protein expression or interfere with immune recognition [68]. It is also associated with various cellular proteins that contribute to viral pathogenesis [69].
3.4.1 Interaction with phospholipid-binding proteins to enhance viral replication
During flavivirus infection, C protein progressively accumulates around lipid droplets (LDs) through a noncanonical function of the COPI system, which provides new ideas for antiviral strategies [70,71,72]. A disordered N-terminal arm of the C protein is involved in specific interactions with host lipid systems, such as LDs and very low-density lipoproteins (VLDL) [73, 74], implicated in viral assembly, maturation and release. The interactions depend on a high concentration of intracellular potassium ions (K+) and are mediated by the surface lipoproteins perilipin 3 α-helix 5 (PLIN3α5) and apolipoprotein Eα-helix 4 (APOEα4). These two proteins are the main ligands of the C protein on the surface of LDs and VLDL [28, 75]. Interestingly, the N-terminal region (first 220 residues) of APOE has multiple motifs that match the C-terminal region (last 220 residues) of PLIN3, and the conserved regions may be primarily involved in specific C protein-LD/VLDL interactions [76]; the L50 and L54 amino acids in the α2 helix of the C protein are crucial for these interactions, which are essential for the viral replication cycle [77]. In the study on the DENV C protein, the authors proved that mutations of these two leucine residues would weaken the interaction between the C protein and LDs. Drugs that disrupt LD biogenesis significantly inhibit viral production during the particle assembly step of the viral replication cycle [78, 79]. Interestingly, the binding of DENV C protein to LDs indicates that a 10 amino acid residue peptide, pep14-23 (14NMLKR18, similar to the importin α self-inhibitory sequence), may be functionally related to the interaction between the C protein and LDs, which may require an α-helical conformation of the C protein. This sequence is a common motif in mosquito-borne flavivirus C proteins. A peptide (pep14-23) designed using the intrinsically disordered N-terminal region (residues 1 to 26) to inhibit the interaction between the DENV C protein and host LDs [78]. In a study on WNV C protein [80], a similar result was observed: pep 14–23 is a potential inhibitor of C protein binding to the host lipid systems, which may be similar to other flaviviruses. The interaction between the C protein and LDs/VLDL is a key step for the viral replication of flaviviruses; therefore, this may provide a novel antiviral strategy.
3.4.2 Interaction with nucleolar proteins to promote nuclear localization
The C protein is transported from the cytoplasm to the nucleus through nuclear pore complexes, which penetrate the double layer of the nuclear envelope. During virus replication, the C protein is located in the nucleus because it interacts with importin in infected cells; however, flavivirus assembly occurs in the cytoplasm [47]. The interaction between the C protein and importin is enhanced by phosphorylation. Hence, dephosphorylation disrupts the interaction, resulting in a decrease in the nuclear localization of the C protein, which provides an opportunity for the assembly of virus nucleocapsid in the cytoplasm. A JEV study [81] demonstrated that the amino acid residues Gly42 and Pro43 are involved in the binding of the C protein to nucleolar phosphoprotein B23, and the binding site is located at amino acid residues 38–77; however, this interaction is not detected in DENV [82]. Interestingly, the mutation of amino acid residues Gly42 and Pro43 of the JEV C protein to Ala influences the nuclear localization of the C protein, which leads to impaired virus replication and pathogenicity [83]. Gly42 and Pro43 are the crucial sites for nuclear localization and are associated with RNA replication, protein synthesis and even propagation in Vero cells [81]. Mori et al. [45] demonstrated that the C protein is present in the nucleolus and cytoplasm of mammalian and insect cell lines after JEV infection or transfection with recombinant plasmids that cause C protein expression. However, the nuclear localization of the JEV C protein has been shown to enhance virus replication.
3.4.3 Interaction with nonsense-mediated mRNA decay (NMD) pathway factors
Zika virus (ZIKV) is an emerging mosquito-borne flavivirus related to DENV and WNV. It can infect human cells in vitro, such as neural progenitor cells (NPCs), and disrupt the nonsense-mediated mRNA decay (NMD) pathway. NMD is a cellular mRNA monitoring mechanism required for the development of normal brain size in mice. Krystal et al. showed that cellular NMD factors, such as the central NMD regulator up-frameshift protein 1 (UPF1), can interact with viral C protein. The expression of C protein post-transcriptionally downregulates the level of UPF1 protein. Cellular fractionation studies have shown that the ZIKV C protein specifically targets nuclear UPF1 and can be degraded by the proteasome [84]. ZIKV uses C protein to reduce UPF1 levels and inhibit the antiviral activity of NMD, which in turn contributes to the development of neuropathology in vivo. Similar phenomena were also found in WNV and DENV. Li et al. also found that the host exon-junction complex (EJC) recycling factor PYM1 can interact with C, thereby interfering with the function and location of the EJC protein. However, EJCs have a role in NMD, and they have antiviral effects in DENV, WNV, and ZIKV by indirectly or directly targeting viral RNA. The EJC protein R8M8A can directly bind to WNV RNA, but depletion of PYM1 attenuates the binding of R8M8A to viral RNA, so WNV infection can segregate PYM1 to protect the RNA from decay [23]. In contrast, PYM1, a capsid-interacting protein, plays a pivotal role in HCV infection [85].
3.4.4 Interaction with hSes3p protein
In the cytoplasm, human Sec 3 (hSec3p) and Jab 1 proteins can interact with the C proteins of DENV and WNV. Confocal experiments showed that hSec3p and flavivirus C protein colocalized in the cytoplasm and perinuclear regions. hSec3p is an extracapsular complex component, and its main function involves secretory pathways and exocytosis. hSec3p has been shown to be a novel chaperone of WNV and DENV C proteins through the proteasome pathway [8]. The SH2 domain-binding motif (last 15 aa) of hSec3p binds to the first 15 amino acids of C; the amino acid residues in positions 14 (WNV) and 13 (DENV) of the C protein are particularly important for this interaction. The protective effect of hSec3p can affect the transcription and translation of viral RNA by chelating elongation factor 1α (EF1α), thereby regulating virus production [25], which can delay flavivirus infection.
3.4.5 Interaction with organelle membranes
C protein is expressed in the ER as a part of the flavivirus polyprotein [86]. A study by Markoff et al. [87] showed that the mature C protein is associated with the ER membrane through the internal hydrophobic region located at the α2/α2’ interface, which is known to be conserved among mosquito- and tick-borne flaviviruses [77, 88, 89]. In the DENV study, the C protein fragment peptide C (pepC) was able to bind negatively charged phospholipid membranes through a charge anchor formed by three positively charged amino acid residues (including Arg-2, Lys-6 and Arg-16) [90]. It interacts with organelle membranes to promote viral replication, virion assembly and viral production. This should be considered in examining the flavivirus life cycle.
3.4.6 Interaction with caprin-1
The expression of the C protein plays an important role in regulating the activity, expression or localization of host molecules [91]. In JEV, YFV and ZIKV, C has been shown to interact with its binding partner caprin-1 to inhibit the formation of stress granules (SGs). The Lys97 and Arg98 amino acid residues in the C protein are important for the interaction with caprin-1, and the mutation of these two amino acids to Ala inhibits the formation of SG and damage virus propagation [92]. However, C protein-mediated suppression of SG formation has not been detected in all flaviviruses; for example, the expression of DENV and WNV C proteins does not significantly block the formation of SG [93]. However, it has been reported that DENV and WNV C proteins interact with other host proteins [94, 95].
3.5 Participation in apoptosis
Another nonstructural function of the multifunctional C protein is involved in apoptosis. The phosphorylation of WNV C protein is very important for its interaction with importin and affects its RNA-binding activity, oligomerization, nuclear localization and apoptosis [46, 96]. In several flaviviruses, C proteins have been shown to be pro-apoptotic. However, in a study by Matt D. Urbanowski, WNV C protein was shown to completely block the apoptosis of infected cells during virus replication. The protective effect is mediated by a phosphatidylinositol 3-kinase (PI3K)-dependent pathway [97]. In the nucleus, nucleolin, the apoptotic protein Daxx [98], core histones (H2A, H2B, H3 and H4), hnRNP-K and Hdm2 (in the case of WNV) interact with the C protein. These interactions affect the induction of apoptosis in the host [96] and regulate transcription, leading to the development of disease. The C proteins of various flaviviruses have pro-apoptotic or anti-apoptotic functions [99, 100].
4 Application of the C protein
Flavivirus C protein is indispensable in virus replication and assembly, can undergo immune escape mutations to efficiently avoid the immune system, and it is an important target of T cells during natural infection [101, 102]. C protein has a specific structure, various functions and biological characteristics, which make it a potentially promising drug target for antiviral agents. In a study of DENV, a novel low-molecular-weight compound, ST-148, was used to interact with the C protein and block its activity, which is required for virus replication [103]. The pep14-23 region of the C protein is the major participant in the interaction with LDs. Therefore, a peptide was designed to bind to LDs and inhibit C protein-LD interactions to affect virus replication. The pep14-23 region is also very important for drug design, but further research is needed. Flavivirus C protein has obvious structural functions in mature virions, especially in the processes of viral encapsidation, because it belongs to the same class of α-helical C proteins as hepadnaviruses and retroviruses. Some studies have successfully used flavivirus C protein in the CTVI strategy against DENV [104, 105] and JEV [24]. In addition to flavivirus, the C protein of classical swine fever virus (CSFV) [24, 106] belonging to the Pestivirus genus has also been used in CTVI. Based on these antiviral mechanisms, CTVI is expected to be applied to more Flaviviridae viruses for targeted antiviral research, such as TMUV, YFV, and the Pegivirus genus. However, there are still some problems to be solved if this strategy is applied to clinical research, such as finding more nucleases that are suitable for CTVI and can be tolerated by host cells without cytotoxicity.
Because envelope domain III (EDIII) contains important linear antigenic epitopes that directly interact with neutralizing antibodies [107] and the C protein is important in the flavivirus life cycle, researchers have fused EDIII of the DENV E protein with the C protein of DENV, leading to the formation of the tetravalent vaccine DIIIC [88, 108, 109]. The results demonstrated that DIIIC can induce cell-mediated immunity and has the ability to protect mice against DENV. The structural flexibility of the C protein has been proven in numerous studies. Schlick et al. [110] used a deletion mutant of the C internal hydrophobic domain and another deletion mutation in the 3′ noncoding region (3′-NCR), which removed a conserved hairpin structure (hairpin II-1 nt 10737–10825) to study attenuated vaccine candidates in WNV in vivo. The vaccine is not pathogenic but can induce protective immunity in mice.
Among all flavivirus proteins, the C protein has the lowest conservation. However, the structural properties of the protein are very similar, and its charge distribution is well conserved, which is conducive to the conservation of its function. Table 1 summarizes the numerous functional applications of flavivirus C proteins.
5 Conclusions
Flavivirus C protein is a multifunctional protein that participates in many aspects of the virus life cycle. The C protein binds to viral RNA to form the nucleocapsid and plays an important role in the process of viral infection, including interactions with cellular proteins and the regulation of cell metabolism, apoptosis and immune responses [111, 112].
As a structural protein, the main function of C involves packaging viral genomic RNA and the formation of the viral core, while anchC may help initiate the formation of the nucleocapsid, including interaction with genomic RNA or oligomerization with other C proteins. The C protein is a highly basic protein rich in positively charged amino acid residues (approximately 26 Arg or Lys residues and only 3 negatively charged residues), which is crucial for its binding and interaction with viral RNA. The C protein is a genomic protective agent that can encapsulate viral RNA through its N- and C-terminal basic amino acid clusters to form the viral nucleocapsid. This indicates that the C protein may be a potentially useful target for the development of antiviral therapy [27, 111, 113]. Flavivirus C protein has highly divergent sequences and different domain organizations. However, all C proteins can promote nucleic acid annealing and enhance hammerhead ribozyme-mediated cleavage. The chaperone activity of the flavivirus C protein is very relevant due to its intrinsic disorder; it has the ability to resist heat denaturation, and boiling C protein for 5 min has no effect on its kinetics [114, 115]. Importantly, the C protein may promote profound structural rearrangement of RNA without consuming ATP. C proteins are conserved in Flaviviridae, including the Hepacivirus (HCV, GBV-B), Pestivirus (bovine viral diarrhoea virus, BVDV), and Flavivirus genera (WNV). In HCV and GBV-B, the N-terminal region of the C protein is a highly basic and flexible RNA-binding domain, while the C-terminal region is a hydrophobic domain [114]. The central hydrophobic region of the C protein may be related to the ER membrane, which is thought to promote nucleocapsid assembly [45]. In addition, the C protein has variable degrees of tolerance to structural changes and can tolerate extensive deletions in its N-/C-terminus, which indicates that the C protein does not require a defined tertiary structure for its function but relies on basic residues to recruit viral RNA. Due to its structural and functional flexibility, the C protein may be a novel and attractive target for the targeted attenuation of flaviviruses. For example, small-molecule inhibitors of the C protein binding sites may be of interest. Recently, it has been proven that the C proteins of ZIKV and YFV can inhibit small RNAs based on the antiviral response [116].
As a “nonstructural” protein, the C protein precursor anchC plays an important role in flavivirus assembly. Prior cleavage at the anchC dibasic site may affect the cleavage mediated by signalase in the ER lumen, and the sequential cleavage of the anchC sequence is considered to be essential for flavivirus production. The mature C protein can regulate virus replication or change the host cell environment [8, 47, 117]. It plays an important role in virus replication through interactions with various host factors [40], such as B23 [118], Jab1, hnRNP K, and hnRNP A2 [119, 120]. The interaction with organelle membranes of the C protein also plays a vital role in the virus life cycle, but the mechanism by which the interactions affect virus propagation during the virus infection cycle has not been reported. Inhibition of the interaction may severely affect virus replication, assembly and release, thereby reducing virus production. Whether or how the conformation of the C protein changes in this process, which region of the four α-helices (α1 to α4) is involved, and how to specifically label the membranes are all worthy of consideration. Understanding and solving these issues will help us to further study antiviral drugs. The interactions between the C protein and host proteins are conserved in DENV, WNV, and ZIKV [23]. The C protein also plays a crucial role in modulating host cell signalling networks by promoting innate immunity or affecting cell apoptosis, which benefits or impedes the flavivirus replication. The C protein also has favourable antigenicity and can induce host cell-mediated immunity [121, 122] and the humoural immune response, so it can be considered a vaccine target candidate. Drugs that interfere with the formation of the C protein or inhibit its conformational changes during the interactions between the C protein and other proteins will affect the process of genome encapsidation and virus release. Overall, the extraordinary functional flexibility of the C protein makes it an attractive target for flavivirus vaccines and vector engineering design, which is very promising and attractive.
Research on the structure and function of flavivirus C proteins provides us with broad application prospects. The existence of a domain with antibacterial activity in the C protein enhances the importance of viral proteins in the drug development platform and poses new challenges to the coevolution of viruses and bacteria. We expect to make increased efforts in the development of vaccines and drugs.
Abbreviations
- C:
-
Capsid
- USUV:
-
Usutu virus
- DENV:
-
Dengue virus
- JEV:
-
Japanese encephalitis virus
- WNV:
-
West Nile virus
- ZIKV:
-
Zika virus
- TBEV:
-
Tick-borne encephalitis virus
- ORF:
-
Open reading frame
- UTR:
-
Untranslated region
- ER:
-
Endoplasmic reticulum
- E:
-
Envelope
- NS:
-
Nonstructural
- anchC:
-
Anchored C
- Ve:
-
Vesicle
- VPs:
-
Vesicle packets
- COPII:
-
Coat protein complex II
- NMR:
-
Nuclear magnetic resonance
- TMUV:
-
Tembusu virus
- NC:
-
Nucleocapsid
- NLSs:
-
Nuclear localization signals
- HCV:
-
Hepatitis C virus
- DCS-PK:
-
Downstream of the 5′ cyclization sequence pseudoknot
- SL6:
-
Stem-loop 6
- CPPs:
-
Cell-penetrating peptides
- YFV:
-
Yellow fever virus
- CTVI:
-
Capsid-targeted antiviral inactivation
- TMD:
-
Transmembrane domain
- HIS:
-
Internal hydrophobic sequence
- LDs:
-
Lipid droplets
- VLDL:
-
Very low-density lipoproteins
- NMD:
-
Nonsense-mediated mRNA decay
- UPF1:
-
Up-frameshift protein 1
- hSec3p:
-
Human Sec 3
- SG:
-
Stress granule
- PI3K:
-
Phosphatidylinositol 3-kinase
- CSFV:
-
Classical swine fever virus
- EDIII:
-
Envelope domain III
References
Chen S, Yang C, Zhang W, Mahalingam S, Wang M, Cheng A (2018) Flaviviridae virus nonstructural proteins 5 and 5A mediate viral immune evasion and are promising targets in drug development. Pharmacol Ther 190:1–14
Sherman KE, Rouster SD, Kong LX, Aliota MT, Blackard JT, Dean GE (2019) Zika virus replication and cytopathic effects in liver cells. PLoS One 14:e0214016
Somvanshi P, Singh V, Seth PK (2009) High throughput prediction and analysis of small interfering RNA from the 5’UTR and capsid genes of flavivirus through in silico strategies. Interdiscip Sci 1:298–302
Shan C, Xie X, Zou J, Züst R, Zhang B, Ambrose R, Mackenzie J, Fink K, Shi PY (2018) Using a virion assembly-defective dengue virus as a vaccine approach. J Virol 92:e01002-18
Lobigs M, Lee E, Ng ML, Pavy M, Lobigs P (2010) A flavivirus signal peptide balances the catalytic activity of two proteases and thereby facilitates virus morphogenesis. Virology 401:80–89
Li G, Poulsen M, Fenyvuesvolgyi C, Yashiroda Y, Yoshida M, Simard JM, Gallo RC, Zhao RY (2017) Characterization of cytopathic factors through genome-wide analysis of the Zika viral proteins in fission yeast. Proc Natl Acad Sci USA 114:E376–E385
Mukhopadhyay S, Kuhn RJ, Rossmann MG (2005) A structural perspective of the flavivirus life cycle. Nat Rev Microbiol 3:13–22
Bhuvanakantham R, Ng ML (2013) West Nile virus and dengue virus capsid protein negates the antiviral activity of human Sec3 protein through the proteasome pathway. Cell Microbiol 15:1688–1706
VanBlargan LA, Davis KA, Dowd KA, Akey DL, Smith JL, Pierson TC (2015) Context-dependent cleavage of the capsid protein by the West Nile virus protease modulates the efficiency of virus assembly. J Virol 89:8632–8642
Pandit PS, Doyle MM, Smart KM, Young CCW, Drape GW, Johnson CK (2018) Predicting wildlife reservoirs and global vulnerability to zoonotic Flaviviruses. Nat Commun 9:5425
Scaturro P, Cortese M, Chatel-Chaix L, Fischl W, Bartenschlager R (2015) Dengue virus non-structural protein 1 modulates infectious particle production via interaction with the structural proteins. PLoS Pathog 11:e1005277
Neufeldt CJ, Cortese M, Scaturro P, Cerikan B, Wideman JG, Tabata K, Moraes T, Oleksiuk O, Pichlmair A, Bartenschlager R (2019) ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat Microbiol 4:2416–2429
Xie X, Zou J, Zhang X, Zhou Y, Routh AL, Kang C, Popov VL, Chen X, Wang Q-Y, Dong H (2019) Dengue NS2A protein orchestrates virus assembly. Cell Host Microbe 26:606-622.e8
Vonderstein K, Nilsson E, Hubel P, Nygard Skalman L, Upadhyay A, Pasto J, Pichlmair A, Lundmark R, Overby AK (2018) Viperin targets flavivirus virulence by inducing assembly of noninfectious capsid particles. J Virol 92:e01751-17
Li T, Zhao Q, Yang X, Chen C, Yang K, Wu C, Zhang T, Duan Y, Xue X, Mi K, Ji X, Wang Z, Yang H (2018) Structural insight into the Zika virus capsid encapsulating the viral genome. Cell Res 28:497–499
Figueira-Mansur J, Aguilera EA, Stoque RM, Ventura GT, Mohana-Borges R (2019) Mutations in the dimer interfaces of the dengue virus capsid protein affect structural stability and impair RNA-capsid interaction. Sci Rep 9:2829
Kiermayr S, Kofler RM, Mandl CW, Messner P, Heinz FX (2004) Isolation of capsid protein dimers from the tick-borne encephalitis flavivirus and in vitro assembly of capsid-like particles. J Virol 78:8078–8084
Clyde K, Barrera J, Harris E (2008) The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 379:314–323
Roby JA, Setoh YX, Hall RA, Khromykh AA (2015) Post-translational regulation and modifications of flavivirus structural proteins. J Gen Virol 96:1551–1569
Mehrbod P, Ande SR, Alizadeh J, Rahimizadeh S, Shariati A, Malek H, Hashemi M, Glover KKM, Sher AA, Coombs KM, Ghavami S (2019) The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence 10:376–413
Freire JM, Santos NC, Veiga AS, Da Poian AT, Castanho MA (2015) Rethinking the capsid proteins of enveloped viruses: multifunctionality from genome packaging to genome transfection. FEBS J 282:2267–2278
Scaturro P, Trist IM, Paul D, Kumar A, Acosta EG, Byrd CM, Jordan R, Brancale A, Bartenschlager R (2014) Characterization of the mode of action of a potent dengue virus capsid inhibitor. J Virol 88:11540–11555
Li M, Johnson JR, Truong B, Kim G, Weinbren N, Dittmar M, Shah PS, Von Dollen J, Newton BW, Jang GM, Krogan NJ, Cherry S, Ramage H (2019) Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat Microbiol 4:985–995
Pang R, He DN, Zhou B, Liu K, Zhao J, Zhang XM, Chen PY (2013) In vitro inhibition of Japanese encephalitis virus replication by capsid-targeted virus inactivation. Antiviral Res 97:369–375
Bhuvanakantham R, Li J, Tan TT, Ng ML (2010) Human Sec3 protein is a novel transcriptional and translational repressor of flavivirus. Cell Microbiol 12:453–472
Jones CT, Ma L, Burgner JW, Groesch TD, Post CB, Kuhn RJ (2003) Flavivirus capsid is a dimeric alpha-helical protein. J Virol 77:7143–7149
Patkar CG, Jones CT, Chang YH, Warrier R, Kuhn RJ (2007) Functional requirements of the yellow fever virus capsid protein. J Virol 81:6471–6481
Carvalho FA, Carneiro FA, Martins IC, Assunção-Miranda I, Faustino AF, Pereira RM, Bozza PT, Castanho MA, Mohana-Borges R, Da Poian AT, Santos NC (2012) Dengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. J Virol 86:2096–2108
Teoh PG, Huang ZS, Pong WL, Chen PC, Wu HN (2014) Maintenance of dimer conformation by the dengue virus core protein α4-α4’ helix pair is critical for nucleocapsid formation and virus production. J Virol 88:7998–8015
Poonsiri T, Wright GSA, Solomon T, Antonyuk SV (2019) Crystal structure of the Japanese encephalitis virus capsid protein. Viruses 11:623
Freire JM, Veiga AS, de la Torre BG, Santos NC, Andreu D, Da Poian AT, Castanho MA (2013) Peptides as models for the structure and function of viral capsid proteins: insights on dengue virus capsid. Biopolymers 100:325–336
Kofler RM, Heinz FX, Mandl CW (2002) Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J Virol 76:3534–3543
Morando MA, Barbosa GM, Cruz-Oliveira C, Da Poian AT, Almeida FCL (2019) Dynamics of Zika virus capsid protein in solution: the properties and exposure of the hydrophobic cleft are controlled by the α-helix 1sequence. Biochemistry 58:2488–2498
Carletti T, Zakaria MK, Faoro V, Reale L, Kazungu Y, Licastro D, Marcello A (2019) Viral priming of cell intrinsic innate antiviral signaling by the unfolded protein response. Nat Commun 10:3889
Ivanyi-Nagy R, Darlix JL (2012) Reprint of: Core protein-mediated 5’-3’ annealing of the West Nile virus genomic RNA in vitro. Virus Res 169:448–457
Zhang X, Xie X, Xia H, Zou J, Huang L, Popov VL, Chen X, Shi PY (2019) Zika virus NS2A-mediated virion assembly. mBio 10:e02375-19
Zheng Y, Kielian M (2013) Imaging of the alphavirus capsid protein during virus replication. J Virol 87:9579–9589
Tan TY, Fibriansah G, Lok SM (2020) Capsid protein is central to the birth of flavivirus particles. PLoS Pathog 16:e1008542
Balinsky CA, Schmeisser H, Ganesan S, Singh K, Pierson TC, Zoon KC (2013) Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol 87:13094–13106
Bhuvanakantham R, Chong MK, Mahlee N (2009) Specific interaction of capsid protein and importin-α/β influences West Nile virus production. Biochem Biophys Res Commun 389:63–69
Idrees S, Ashfaq UA (2012) A brief review on dengue molecular virology, diagnosis, treatment and prevalence in Pakistan. Genet Vaccines Ther 10:6
Wang SH, Syu WJ, Huang KJ, Lei HY, Yao CW, King CC, Hu ST (2002) Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J Gen Virol 83:3093–3102
Netsawang J, Noisakran S, Puttikhunt C, Kasinrerk W, Wongwiwat W, Malasit P, Yenchitsomanus PT, Limjindaporn T (2010) Nuclear localization of dengue virus capsid protein is required for DAXX interaction and apoptosis. Virus Res 147:275–283
Samsa MM, Mondotte JA, Caramelo JJ, Gamarnik AV (2012) Uncoupling cis-Acting RNA elements from coding sequences revealed a requirement of the N-terminal region of dengue virus capsid protein in virus particle formation. J Virol 86:1046–1058
Mori Y, Okabayashi T, Yamashita T, Zhao Z, Wakita T, Yasui K, Hasebe F, Tadano M, Konishi E, Moriishi K, Matsuura Y (2005) Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J Virol 79:3448–3458
Cheong YK, Ng ML (2011) Dephosphorylation of West Nile virus capsid protein enhances the processes of nucleocapsid assembly. Microbes Infect 13:76–84
Bhuvanakantham R, Cheong YK, Ng ML (2010) West Nile virus capsid protein interaction with importin and HDM2 protein is regulated by protein kinase C-mediated phosphorylation. Microbes Infect 12:615–625
Baker C, Xie X, Zou J, Muruato A, Fink K, Shi PY (2020) Using recombination-dependent lethal mutations to stabilize reporter flaviviruses for rapid serodiagnosis and drug discovery. EBioMedicine 57:102838
Schlick P, Taucher C, Schittl B, Tran JL, Kofler RM, Schueler W, von Gabain A, Meinke A, Mandl CW (2009) Helices alpha2 and alpha3 of West Nile virus capsid protein are dispensable for assembly of infectious virions. J Virol 83:5581–5591
Schrauf S, Schlick P, Skern T, Mandl CW (2008) Functional analysis of potential carboxy-terminal cleavage sites of tick-borne encephalitis virus capsid protein. J Virol 82:2218–2229
Tsetsarkin KA, Liu G, Shen K, Pletnev AG (2016) Kissing-loop interaction between 5’ and 3’ ends of tick-borne Langat virus genome “bridges the gap” between mosquito- and tick-borne flaviviruses in mechanisms of viral RNA cyclization: applications for virus attenuation and vaccine development. Nucleic Acids Res 44:3330–3350
de Borba L, Villordo SM, Iglesias NG, Filomatori CV, Gebhard LG, Gamarnik AV (2015) Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J Virol 89:3430–3437
Liu ZY, Li XF, Jiang T, Deng YQ, Zhao H, Wang HJ, Ye Q, Zhu SY, Qiu Y, Zhou X, Qin ED, Qin CF (2013) Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. J Virol 87:6804–6818
Tuplin A, Evans DJ, Buckley A, Jones IM, Gould EA, Gritsun TS (2011) Replication enhancer elements within the open reading frame of tick-borne encephalitis virus and their evolution within the Flavivirus genus. Nucleic Acids Res 39:7034–7048
Xie X, Zou J, Zhang X, Zhou Y, Routh AL, Kang C, Popov VL, Chen X, Wang QY, Dong H, Shi PY (2019) Dengue NS2A protein orchestrates virus assembly. Cell Host Microbe 26:606–622
Qi X, Droste T, Kao CC (2011) Cell-penetrating peptides derived from viral capsid proteins. Mol Plant Microbe Interact 24:25–36
Freire JM, Veiga AS, Conceição TM, Kowalczyk W, Mohana-Borges R, Andreu D, Santos NC, Da Poian AT, Castanho MA (2013) Intracellular nucleic acid delivery by the supercharged dengue virus capsid protein. PLoS One 8:e81450
Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB (2004) Solution structure of dengue virus capsid protein reveals another fold. Proc Natl Acad Sci USA 101:3414–3419
Freire JM, Veiga AS, Rego de Figueiredo I, de la Torre BG, Santos NC, Andreu D, Da Poian AT, Castanho MA (2014) Nucleic acid delivery by cell penetrating peptides derived from dengue virus capsid protein: design and mechanism of action. FEBS J 281:191–215
Samuel GH, Wiley MR, Badawi A, Adelman ZN, Myles KM (2016) Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc Natl Acad Sci USA 113:13863–13868
Alcaraz-Estrada SL, Del Angel R, Padmanabhan R (2014) Construction of self-replicating subgenomic dengue virus 4 (DENV4) replicon. Methods Mol Biol 1138:131–150
Natsoulis G, Boeke JD (1991) New antiviral strategy using capsid-nuclease fusion proteins. Nature 352:632–635
Zhang X, Jia R, Zhou J, Wang M, Yin Z, Cheng A (2016) Capsid-targeted viral inactivation: a novel tactic for inhibiting replication in viral infections. Viruses 8:258
Rana J, Slon Campos JL, Leccese G, Francolini M, Bestagno M, Poggianella M, Burrone OR (2018) Role of capsid anchor in the morphogenesis of Zika virus. J Virol 92:e01174-18
Perry JW, Chen Y, Speliotes E, Tai AW (2018) Functional analysis of the Dengue virus genome using an insertional mutagenesis screen. J Virol 92:e02085-17
Gillespie LK, Hoenen A, Morgan G, Mackenzie JM (2010) The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol 84:10438–10447
Kofler RM, Leitner A, O’Riordain G, Heinz FX, Mandl CW (2003) Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J Virol 77:443–451
Klumpp K, Crépin T (2014) Capsid proteins of enveloped viruses as antiviral drug targets. Curr Opin Virol 5:63–71
Kumar R, Singh N, Abdin MZ, Patel AH, Medigeshi GR (2017) Dengue virus capsid interacts with DDX3X-A potential mechanism for suppression of antiviral functions in Dengue infection. Front Cell Infect Microbiol 7:542
Villareal VA, Rodgers MA, Costello DA, Yang PL (2015) Targeting host lipid synthesis and metabolism to inhibit dengue and hepatitis C viruses. Antiviral Res 124:110–121
Taguwa S, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, Andino R, Fernandez-Sesma A, Frydman J (2015) Defining Hsp70 subnetworks in Dengue virus replication reveals key vulnerability in Flavivirus infection. Cell 163:1108–1123
Taguwa S, Yeh MT, Rainbolt TK, Nayak A, Shao H, Gestwicki JE, Andino R, Frydman J (2019) Zika virus dependence on host Hsp70 provides a protective strategy against infection and disease. Cell Rep 26:906–920
Guevara J Jr, Romo J Jr, McWhorter T, Guevara NV (2015) Analogs of LDL receptor ligand motifs in dengue envelope and capsid proteins as potential codes for cell entry. J Viruses 2015:646303
Faustino AF, Carvalho FA, Martins IC, Castanho MA, Mohana-Borges R, Almeida FC, Da Poian AT, Santos NC (2014) Dengue virus capsid protein interacts specifically with very low-density lipoproteins. Nanomedicine 10:247–255
Martins AS, Carvalho FA, Faustino AF, Martins IC, Santos NC (2019) West Nile virus capsid protein interacts with biologically relevant host lipid systems. Front Cell Infect Microbiol 9:8
Faustino AF, Martins IC, Carvalho FA, Castanho MA, Maurer-Stroh S, Santos NC (2015) Understanding Dengue virus capsid protein interaction with key biological targets. Sci Rep 5:10592
Samsa MM, Mondotte JA, Iglesias NG, Assunção-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV (2009) Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog 5:e1000632
Faustino AF, Guerra GM, Huber RG, Hollmann A, Domingues MM, Barbosa GM, Enguita FJ, Bond PJ, Castanho MARB, Poian ATD (2014) Understanding Dengue virus capsid protein disordered N-terminus and pep14-23-based inhibition. ACS Chem Biol 10:517–526
Martins AS, Martins IC, Santos NC (2018) Methods for lipid droplet biophysical characterization in Flaviviridae infections. Front Microbiol 9:1951
Jabłońska E, Reszka E (2017) Selenum and epigenetics in cancer: focus on DNA methylation. Adv Cancer Res 136:193–234
Tsuda Y, Mori Y, Abe T, Yamashita T, Okamoto T, Ichimura T, Moriishi K, Matsuura Y (2006) Nucleolar protein B23 interacts with Japanese encephalitis virus core protein and participates in viral replication. Microbiol Immunol 50:225–234
Iglesias NG, Mondotte JA, Byk LA, De Maio FA, Samsa MM, Alvarez C, Gamarnik AV (2015) Dengue virus uses a non-canonical function of the host GBF1-Arf-COPI system for capsid protein accumulation on lipid droplets. Traffic 16:962–977
Wulan WN, Heydet D, Walker EJ, Gahan ME, Ghildyal R (2015) Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front Microbiol 6:553–562
Fontaine KA, Leon KE, Khalid MM, Tomar S, Jimenez-Morales D, Dunlap M, Kaye JA, Shah PS, Finkbeiner S, Krogan NJ (2018) The cellular NMD pathway restricts Zika virus infection and is targeted by the viral capsid protein. MBio 9:e02126-18
Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, Ott M (2015) A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell 57:329–340
Roby JA, Hall RA, Khromykh AA (2013) West Nile virus genome with glycosylated envelope protein and deletion of alpha helices 1, 2, and 4 in the capsid protein is noninfectious and efficiently secretes subviral particles. J Virol 87:13063–13069
Markoff L, Falgout B, Chang A (1997) A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology 233:105–117
Lazo L, Valdes I, Guillén G, Hermida L, Gil L (2019) Aiming at the heart: the capsid protein of dengue virus as a vaccine candidate. Expert Rev Vaccines 18:161–173
Nemésio H, Palomares-Jerez MF, Villalaín J (2013) Hydrophobic segment of dengue virus C protein. Interaction with model membranes. Mol Membr Biol 30:273–287
Fajardo-Sánchez E, Galiano V, Villalaín J (2017) Molecular dynamics study of the membrane interaction of a membranotropic dengue virus C protein-derived peptide. J Biomol Struct Dyn 35:1283–1294
Basu M, Courtney SC, Brinton MA (2017) Arsenite-induced stress granule formation is inhibited by elevated levels of reduced glutathione in West Nile virus-infected cells. PLoS Pathog 13:e1006240
Katoh H, Okamoto T, Fukuhara T, Kambara H, Morita E, Mori Y, Kamitani W, Matsuura Y (2013) Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with Caprin-1 and facilitates viral propagation. J Virol 87:489–502
Hou S, Kumar A, Xu Z, Airo AM, Stryapunina I, Wong CP, Branton W, Tchesnokov E, Götte M, Power C, Hobman TC (2017) Zika virus hijacks stress granule proteins and modulates the host stress response. J Virol 91:e00474-17
Xu Z, Anderson R, Hobman TC (2011) The capsid-binding nucleolar helicase DDX56 is important for infectivity of West Nile virus. J Virol 85:5571–5580
Xu Z, Hobman TC (2012) The helicase activity of DDX56 is required for its role in assembly of infectious West Nile virus particles. Virology 433:226–235
Yang MR, Lee SR, Oh W, Lee EW, Yeh JY, Nah JJ, Joo YS, Shin J, Lee HW, Pyo S, Song J (2008) West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell Microbiol 10:165–176
Urbanowski MD, Hobman TC (2013) The West Nile virus capsid protein blocks apoptosis through a phosphatidylinositol 3-kinase-dependent mechanism. J Virol 87:872–881
Limjindaporn T, Netsawang J, Noisakran S, Thiemmeca S, Wongwiwat W, Sudsaward S, Avirutnan P, Puttikhunt C, Kasinrerk W, Sriburi R, Sittisombut N, Yenchitsomanus PT, Malasit P (2007) Sensitization to Fas-mediated apoptosis by dengue virus capsid protein. Biochem Biophys Res Commun 362:334–339
Slomnicki LP, Chung DH, Parker A, Hermann T, Boyd NL, Hetman M (2017) Ribosomal stress and Tp53-mediated neuronal apoptosis in response to capsid protein of the Zika virus. Sci Rep 7:16652
Yang JS, Ramanathan MP, Muthumani K, Choo AY, Jin SH, Yu QC, Hwang DS, Choo DK, Lee MD, Dang K, Tang W, Kim JJ, Weiner DB (2002) Induction of inflammation by West Nile virus capsid through the caspase-9 apoptotic pathway. Emerg Infect Dis 8:1379–1384
Lim MQ, Kumaran EAP, Tan HC, Lye DC, Leo YS, Ooi EE, MacAry PA, Bertoletti A, Rivino L (2018) Cross-reactivity and anti-viral function of Dengue capsid and NS3-specific memory T cells toward Zika virus. Front Immunol 9:2225–2238
Shi X, Hu J, Guo J, Wu C, Xiong S, Dong C (2019) A vesicular stomatitis virus-based vaccine carrying Zika virus capsid protein protects mice from viral infection. Virol Sin 34:106–110
Byrd CM, Dongcheng D, Grosenbach DW, Aklile B, Jones KF, Cardwell KB, Christine S, Wineinger KA, Page JM, Chris H (2013) A novel inhibitor of dengue virus replication that targets the capsid protein. Antimicrob Agents Chemother 57:15–25
Qin CF, Jiang T, Chen SP, Yu M, Qin ED (2005) Capsid-targeted viral inactivation for dengue virus infection. Acta Microbiol Sin 45:111–115
Qin CF, Qin ED (2004) Development of cell lines stably expressing staphylococcal nuclease fused to dengue 2 virus capsid protein for CTVI. Acta Biochim Biophys Sin 36:577–582
Zhou B, Liu K, Wei JC, Mao X, Chen PY (2010) Inhibition of replication of classical swine fever virus in a stable cell line by the viral capsid and Staphylococcus aureus nuclease fusion protein. J Virol Methods 167:79–83
Izquierdo A, García A, Lazo L, Gil L, Marcos E, Alvarez M, Valdés I, Hermida L, Guillén G, Guzmán MG (2014) A tetravalent dengue vaccine containing a mix of domain III-P64k and domain III-capsid proteins induces a protective response in mice. Arch Virol 159:2597–2604
Valdés I, Marcos E, Suzarte E, Pérez Y, Brown E, Lazo L, Cobas K, Yaugel M, Rodríguez Y, Gil L, Guillén G, Hermida L (2017) A dose-response study in mice of a tetravalent vaccine candidate composed of domain III-capsid proteins from dengue viruses. Arch Virol 162:2247–2256
Izquierdo A, Valdés I, Gil L, Hermida L, Gutiérrez S, García A, Bernardo L, Pavón A, Guillén G, Guzmán MG (2012) Serotype specificity of recombinant fusion protein containing domain III and capsid protein of dengue virus 2. Antiviral Res 95:1–8
Schlick P, Kofler RM, Schittl B, Taucher C, Nagy E, Meinke A, Mandl CW (2010) Characterization of West Nile virus live vaccine candidates attenuated by capsid deletion mutations. Vaccine 28:5903–5909
Tan TY, Fibriansah G, Kostyuchenko VA, Ng TS, Lim XX, Zhang S, Lim XN, Wang J, Shi J, Morais MC, Corti D, Lok SM (2020) Capsid protein structure in Zika virus reveals the flavivirus assembly process. Nat Commun 11:895
Shang Z, Song H, Shi Y, Qi J, Gao GF (2018) Crystal structure of the capsid protein from Zika virus. J Mol Biol 430:948–962
Byk LA, Iglesias NG, De Maio FA, Gebhard LG, Rossi M, Gamarnik AV (2016) Dengue virus genome uncoating requires ubiquitination. mBio 7:e00804-16
Ivanyi-Nagy R, Lavergne JP, Gabus C, Ficheux D, Darlix JL (2008) RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res 36:712–725
Pong WL, Huang ZS, Teoh PG, Wang CC, Wu HN (2011) RNA binding property and RNA chaperone activity of dengue virus core protein and other viral RNA-interacting proteins. FEBS Lett 585:2575–2581
Varjak M, Donald CL, Mottram TJ, Sreenu VB, Merits A, Maringer K, Schnettler E, Kohl A (2017) Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. PLoS Negl Trop Dis 11:e0006010
Mori Y, Yamashita T, Tanaka Y, Tsuda Y, Abe T, Moriishi K, Matsuura Y (2007) Processing of capsid protein by cathepsin L plays a crucial role in replication of Japanese encephalitis virus in neural and macrophage cells. J Virol 81:8477–8487
Tiwary AK, Cecilia D (2017) Kinetics of the association of dengue virus capsid protein with the granular component of nucleolus. Virology 502:48–55
Barrows NJ, Campos RK, Liao KC, Prasanth KR, Soto-Acosta R, Yeh SC, Schott-Lerner G, Pompon J, Sessions OM, Bradrick SS, Garcia-Blanco MA (2018) Biochemistry and molecular biology of Flaviviruses. Chem Rev 118:4448–4482
Ishida K, Goto S, Ishimura M, Amanuma M, Hara Y, Suzuki R, Katoh K, Morita E (2019) Functional correlation between subcellular localizations of Japanese Encephalitis virus capsid protein and virus production. J Virol 93:e00612-19
Gil L, Cobas K, Lazo L, Marcos E, Hernández L, Suzarte E, Izquierdo A, Valdés I, Blanco A, Puentes P, Romero Y, Pérez Y, Guzmán MG, Guillén G, Hermida L (2016) A tetravalent formulation based on recombinant nucleocapsid-like particles from dengue viruses induces a functional immune response in mice and monkeys. J Immunol 197:3597–3606
Weiskopf D, Cerpas C, Angelo MA, Bangs DJ, Sidney J, Paul S, Peters B, Sanches FP, Silvera CG, Costa PR, Kallas EG, Gresh L, de Silva AD, Balmaseda A, Harris E, Sette A (2015) Human CD8+ T-cell responses against the 4 dengue virus serotypes are associated with distinct patterns of protein targets. J Infect Dis 212:1743–1751
Gil L, Marcos E, Izquierdo A, Lazo L, Valdés I, Ambala P, Ochola L, Hitler R, Suzarte E, Álvarez M, Kimiti P, Ndung’u J, Kariuki T, Guzmán MG, Guillén G, Hermida L (2015) The protein DIIIC-2, aggregated with a specific oligodeoxynucleotide and adjuvanted in alum, protects mice and monkeys against DENV-2. Immunol Cell Biol 93:57–66
Suzarte E, Gil L, Valdés I, Marcos E, Lazo L, Izquierdo A, García A, López L, Álvarez M, Pérez Y, Castro J, Romero Y, Guzmán MG, Guillén G, Hermida L (2015) A novel tetravalent formulation combining the four aggregated domain III-capsid proteins from dengue viruses induces a functional immune response in mice and monkeys. Int Immunol 27:367–379
Gil L, Lazo L, Valdes I, Suzarte E, Yen P, Ramirez R, Alvarez M, Dung LT, Cobas K, Marcos E, Perez Y, Guzman MG, Ngyen DH, Guillen G, Hermida L (2017) The tetravalent formulation of domain III-capsid proteins recalls memory B- and T-cell responses induced in monkeys by an experimental dengue virus infection. Clin Transl Immunol 6:e148
Valdés I, Bernardo L, Gil L, Pavón A, Lazo L, López C, Romero Y, Menendez I, Falcón V, Betancourt L, Martín J, Chinea G, Silva R, Guzmán MG, Guillén G, Hermida L (2009) A novel fusion protein domain III-capsid from dengue-2, in a highly aggregated form, induces a functional immune response and protection in mice. Virology 394:249–258
Gil L, Izquierdo A, Lazo L, Valdés I, Ambala P, Ochola L, Marcos E, Suzarte E, Kariuki T, Guzmán G (2014) Capsid protein: Evidences about the partial protective role of neutralizing antibody-independent immunity against dengue in monkeys. Virology 456–457:70–76
Kumar AS, Reddy GE, Rajmane Y, Nair S, Pai Kamath S, Sreejesh G, Basha K, Chile S, Ray K, Nelly V, Khadpe N, Kasturi R, Ramana V (2015) siRNAs encapsulated in recombinant capsid protein derived from Dengue serotype 2 virus inhibits the four serotypes of the virus and proliferation of cancer cells. J Biotechnol 193:23–33
Huang J, Shen H, Jia R, Wang M, Chen S, Zhu D, Liu M, Zhao X, Yang Q, Wu Y (2018) Oral vaccination with a DNA vaccine encoding capsid protein of duck tembusu virus induces protection immunity. Viruses 10:180
Zhang X, Jia R, Pan Y, Wang M, Chen S, Zhu D, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yin Z, Jing B, Huang J, Tian B, Pan L, Yu Y, Ur Rehman M, Cheng A (2019) Therapeutic effects of duck Tembusu virus capsid protein fused with staphylococcal nuclease protein to target Tembusu infection in vitro. Vet Microbiol 235:295–300
Garg H, Sedano M, Plata G, Punke EB, Joshi A (2017) Development of virus-like-particle vaccine and reporter assay for Zika virus. J Virol 91:e00834-17
Xie X, Kum DB, Xia H, Luo H, Shan C, Zou J, Muruato AE, Medeiros DBA, Nunes BTD, Dallmeier K, Rossi SL, Weaver SC, Neyts J, Wang T, Vasconcelos PFC, Shi PY (2018) A single-dose live-attenuated Zika virus vaccine with controlled infection rounds that protects against vertical transmission. Cell Host Microbe 24:487–499
Acknowledgements
We apologize to the authors of articles reporting relevant research that were not cited in this manuscript due to limited space.
Funding
This work was supported by the National Natural Science Foundation of China (31872475), Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (CARS-SVDIP), and the China Agricultural Research System (CARS-42-17).
Author information
Authors and Affiliations
Contributions
XZ conceived and wrote the paper, and YZ conceived and revised the paper. RJ and ZY contributed to the English proofreading. MW and AC were responsible for revising the manuscript critically for expert content. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, X., Zhang, Y., Jia, R. et al. Structure and function of capsid protein in flavivirus infection and its applications in the development of vaccines and therapeutics. Vet Res 52, 98 (2021). https://doi.org/10.1186/s13567-021-00966-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-021-00966-2