Abstract
Background
This study investigated the role of Forkhead box Q1 (FOXQ1) in the osteogenic differentiation of bone mesenchymal stem cells.
Methods
Mouse bone mesenchymal stem cells (mBMSCs) were transfected with lentivirus to generate Foxq1-overexpressing mBMSCs, Foxq1-suppressed mBMSCs, and mBMSC controls. The activity of osteogenic differentiation was evaluated with alizarin red staining, alkaline phosphatase activity assay, and RT-qPCR. Wnt/β-catenin signaling activities were compared among groups by TOPFlash/FOPFlash assay, immunofluorescence staining, and western blot assay of beta-catenin (CTNNB1). Coimmunoprecipitation mass spectrometry was also carried out to identify proteins binding with FOXQ1.
Results
Our data showed that FOXQ1 expression was positively correlated with the osteogenic differentiation of the mBMSCs. FOXQ1 also promoted the nuclear translocation of CTNNB1 in the mBMSCs, enhancing Wnt/β-catenin signaling, which was also shown to be essential for the osteogenic differentiation-promoting effect of FOXQ1 in the mBMSCs. Annexin A2 (ANXA2) was bound with FOXQ1, and its depletion reversed the promoting effect of FOXQ1 on Wnt/β-catenin signaling.
Conclusion
These results showed that FOXQ1 binds with ANXA2, promoting Wnt/β-catenin signaling in bone mesenchymal stem cells, which subsequently promotes osteogenic differentiation.
Similar content being viewed by others
Background
Mesenchymal stem cells constitute a group of multipotent cells capable of differentiating into various types of cells, including osteoblasts, adipocytes, and various other types of cells. This process is modulated by various signaling pathways, including BMP [1], TGF-β [2], and the Wnt/β-catenin [3] pathway. The Wnt/β-catenin signaling pathway plays an important role in mesenchymal stem cell stimulation and differentiation regulation [4, 5]. The main event in Wnt/β-catenin signaling involves the stabilization and nuclear translocation of beta-catenin (CTNNB1), which then forms a complex with TCF/LEF and initiates downstream gene transcription [6]. Similar to other major signaling pathways, Wnt/β-catenin is regulated by multiple factors [7].
Forkhead box Q1 (FOXQ1, also known as HFH1) is a member of the forkhead box (FOX) family of proteins. It was first identified as a regulator of hair follicle development [8]. Later identified as an oncogene, FOXQ1 is highly expressed in colorectal cancer, breast cancer, liver cancer, and various other cancers [9], and multiple studies have demonstrated a close relationship between FOXQ1 and Wnt/β-catenin in cancer cells [10, 11]. FOXQ1 also regulates various physiological processes, including the survival [12] and proliferation of stem cells [13]; however, its role in osteogenic differentiation remains to be elucidated.
In the current study, we aimed to investigate the influence of FOXQ1 on the osteogenic differentiation of mesenchymal stem cells and to elucidate the underlying molecular mechanisms. Our findings suggested that FOXQ1 promotes osteogenic differentiation of mouse bone mesenchymal stem cells via the Wnt/β-catenin signaling pathway.
Methods
Animal study and ethical approval of the protocol
To obtain mouse embryo and alveolar bone tissue for histological evaluation, pregnant Chinese Kunming (KM) mice (4 weeks old) and Chinese Kunming mice (7 days old) were purchased from Sun Yat-sen University. Chinese Kunming mice (7 days old) came from the same pregnant mouse to minimize the genetic difference between groups. The study protocol was approved by the Ethics Committee of the Hospital of Stomatology, Sun Yat-sen University (ERC-2013-15; Guangzhou, China).
Tissue preparation and histology evaluation
To observe FOXQ1 expression in alveolar bone tissue, a pregnant KM mouse was sacrificed to obtain 3 mouse embryos at embryonic day 15.5 (E15.5), and 3 KM mice were sacrificed at each time point (postnatal day 7 (P7) and postnatal day 11 (P11)), whose mandibles were isolated surgically. The whole embryos of E15.5 mouse and the mandibles from P7 and P11 mice were fixed with 4% paraformaldehyde at room temperature for 72 h. Then, the samples were dehydrated with graded solutions of alcohol and embedded. Anti-FOXQ1 polyclonal antibody (5 μg/mL; MBS9408074; My BioSource, Inc., San Diego, USA) was used as the primary antibody.
Immunofluorescence analyses were carried out to evaluate the transnucleation of CTNNB1. Cells were incubated overnight with anti-CTNNB1 polyclonal antibody (5 μg/mL; ab2365; Abcam, Cambridge, UK) at 4 °C. Then, the sections were incubated with secondary antibody (1:1000 dilution; A-21206; Invitrogen, CA, USA) for 1 h in a dark chamber. Finally, the sections were counterstained with 4′6-diamidino-2-phenylindole (DAPI; 0.5 μg/mL; Thermo Fisher Scientific, MA, USA) for 15 min for nuclear labeling.
Cell culture
Commercially available mouse bone mesenchymal stem cells (mBMSCs) derived from the bone marrow of Balb/c mice (MUCMX-01001; Cyagen Biosciences; Guangdong, China) were purchased and cultured with alpha-modified Eagle medium (⍺-MEM; Life Technologies, CA, USA) supplemented with 10% fetal bovine serum (FBS, Life Technologies, CA, USA), 100 U/mL penicillin (Sigma, MO, USA), and 100 mg/mL streptomycin (Sigma, MO, USA). After the 3rd passage, the cells were used for experiments.
FOXQ1 expression manipulation
Full-length Foxq1 cDNA was amplified with Flag-tagged primers from total RNA and then cloned into a pCDNA3.1 vector (V79020, Thermo Fisher Scientific, MA, USA) to produce the bait, Flag-tagged FOXQ1 protein, for the coimmunoprecipitation study. Full-length Foxq1 cDNA was also cloned from total RNA and inserted into a pGLV5 vector (GenePharma, Shanghai, China). PGLV3 lentivirus containing the Foxq1 shRNA sequence and lentivirus particles with an empty pGLV3 vector and with a pGLV5 plasmid were purchased from GenePharma (GenePharma, Shanghai, China). MBMSCs were then transfected with lentiviral particles containing Foxq1-sh pGLV3, Foxq1-overexpressing pGLV5, an empty pGLV3, or an empty pGLV5 vector, creating 4 groups of cells denoted as Foxq1-sh mBMSCs, Foxq1-over mBMSCs, lv3 mBMSCs, and lv5 mBMSCs, respectively. Real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR) and western blot assay were carried out to assess Foxq1 expression in each group of mBMSCs.
Osteogenic differentiation, alizarin red staining, and alkaline phosphatase activity assay
Mouse bone mesenchymal stem cells were seeded into 6-well plates at a density of 1.0 × 106 cells per well. An osteogenic induction medium was prepared according to previous studies [14]. The cells were cultured in the induction medium for 14 days, during which period the medium was changed every 3 days, and then evaluated.
Alizarin red staining (ARS) was conducted to visualize the mineral deposition in each group after osteogenic induction. The cells were first fixed with cold methanol at room temperature for 10 min, rinsed twice with deionized water, and stained with alizarin red (10 μL/mL, 130-22-3; Sigma, MO, USA) at room temperature for 30 min. Then, the excess dye was removed with deionized water. For quantification of ARS, stain was desorbed with 10% cetylpyridinium chloride (CPC) in PBS, pH 7.0, for 15 min at room temperature. Then, ARS concentration was determined by absorbance at 560 nm with a spectrometer.
Alkaline phosphatase activity assay was also carried out to evaluate the osteogenic differentiation of mBMSCs from 4 groups. Cultured in the osteogenic induction medium for 7 days, 4 groups of cells were then lysed with 200 μL 1% Triton X-100. Cell lysates were then measured for alkaline phosphatase (ALP) activity with ALP measuring kit (P0321S; Beyotime, Shanghai, China). The ALP activity was measured at 405-nm wavelength with a spectrometer.
Quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 4 groups of mBMSCs using TRIzol (Invitrogen, NY, USA) according to the manufacturer’s protocol. Complimentary DNA synthesis was performed with random 6-mer primers using a PrimeScript 1st Strand cDNA synthesis kit (6110A; TaKaRa Bio, Shiga, Japan). Messenger RNA expression was measured by RT-qPCR using SYBR® Green.
Various markers indicating activities of mesenchymal stem cell osteogenic differentiation were evaluated. Also, to investigate the effect of counterregulating Wnt signaling on FOXQ1’s osteogenic differentiation-promoting action in mBMSCs, Dickkopf WNT signaling pathway inhibitor 1 (DKK1, 200 ng/mL; 5897-DK; R & D Systems, Minneapolis, USA) was used to suppress Wnt signaling in the Foxq1-overexpressing mBMSCs, while 6-bromoindirubin-3′-oxime (6BIO, 2 μg/mL; 3194; R & D Systems, Minneapolis, USA) was used to activate Wnt signaling in the Foxq1-sh mBMSCs. The mRNA levels of Wnt1, Wnt3, and Wnt10a were then evaluated with RT-qPCR. The relative fold change for the expression of the target gene was normalized to the level of glyceraldehyde-3-phosphate dehydrogenase (Gapdh). The primers employed are listed in Table 1.
Western blot analysis
Foxq1-sh mBMSCs, Foxq1-over mBMSCs, lv3 mBMSCs, and lv5 mBMSCs were seeded at a density of 1 × 106 cells per well. Total proteins were extracted using RIPA buffer following the manufacturer’s protocol. Nuclear protein was isolated with a nuclear protein extraction kit according to the manufacturer’s instructions (78833; Thermo Fisher Scientific, MA, USA). The primary antibody was anti-CTNNB1 (1 μg/mL; ab2365; Abcam, Cambridge, UK) and anti-FOXQ1 (0.5 μg/mL; sc-166265; Santa Cruz Biotechnology, Dallas, USA). Anti-THOC1 (2 μg/mL; ab487; Abcam, Cambridge, UK), anti-EEF1A1 (1 μl/mL; ab140632; Abcam, Cambridge, UK), and anti-GAPDH (1 μl/mL; ab9485; Abcam, Cambridge, UK) were used as controls for the nuclear proteins, cytoplasmic proteins, and total proteins, respectively. All assays were performed in triplicate. Quantitative analysis of western blot assays was carried out with ImageJ software [15].
TOPFlash/FOPFlash reporter assay
To assess the influence of FOXQ1 levels on Wnt/β-catenin signaling activities, a TOPFlash/FOPFlash reporter assay was performed. Cells from the Foxq1-sh mBMSC, Foxq1-over mBMSC, lv3 mBMSC, and lv5 mBMSC groups and Foxq1-over+siAnxa2 mBMSCs were seeded on 96-well plates at a density of 4 × 103 cells per well. Then, they were transiently transfected with TOPFlash or FOPFlash luciferase reporter plasmid (17-285; Millipore Sigma; MA, USA) according to the manufacturer’s protocol. The firefly luciferase activity level was normalized against the Renilla luciferase activity level. The fold increase indicating the TOPFlash activity compared to the FOPFlash is reported.
Coimmunoprecipitation
To investigate the potential mechanism of the interaction between FOXQ1 and Wnt/β-catenin and subsequent signaling, coimmunoprecipitation was performed and the resultant data analyzed. Foxq1-overexpressing mBMSCs and lv5 mBMSCs were transfected with the Flag-tagged Foxq1 pCDNA3.1 vector. Total protein was obtained from cell lysates from each group of cells. Coimmunoprecipitation (co-Ip) was carried out with a Pierce co-IP kit (26149; ThermoFisher Scientific, MA, USA) following the manufacturer’s protocol. In brief, 10 μg of anti-FLAG antibody (F3165, Sigma, MA, USA) was first incubated with coupling resin. The antibody-conjugated resin was then incubated overnight with 200 μL mBMSC total protein lysates at 4 °C. Then, the resin was washed, and protein complexes bound to the anti-FLAG antibody were eluted. A western blot analysis was subsequently performed as described above to confirm that the anti-FLAG antibody pulled down FOXQ1 proteins.
Liquid chromatography–mass spectrometry and data analysis
Liquid chromatography–mass spectrometry (LC-MS) analysis was conducted with a NanoLC 400 system, and a TripleTOF 5600-Plus (AB Sciex, Toronto, Canada) system was used for the mass spectrometry (MS) analysis. ProteinPilot software (AB Sciex, Toronto, Canada) was used to analyze data from the TripleTOF 5600-Plus and identified proteins bound to FOXQ1.
The potential interactions among identified proteins were evaluated with the STRING pathway database [16]. Specifically, we tried to identify proteins associated with the Wnt/β-catenin pathway, and the proteins that were identified through the use of the STRING database were then ranked by their percentage of coverage in the LC-MS/MS results.
Statistical analysis
Upon confirmation of a normal distribution of data, all the quantitative data were subjected to Student’s t tests (comparison between two groups) or one-way ANOVA, and Dunnett’s test was used as a post hoc test (comparison between 3 or more groups). P < 0.05 was considered significant. Statistical analyses were carried out using R 3.4.2 (R Foundation, Vienna, Austria).
Result
FOXQ1 expression was associated with osteogenic differentiation of mesenchymal cells in alveolar bone tissue
Immunohistochemical staining demonstrated a high level of FOXQ1 in alveolar bone tissue from the E15.5 mouse embryos (Fig. 1A (a1, a2)). The FOXQ1 protein level was also high in the bone tissue of the P7 mice (Fig. 1A (b1, b2)) and the P11 mice (Fig. 1A (c1, c2)) but was lower than that in the E15.5. Immunofluorescence demonstrated a high level of FOXQ1 protein in mouse bone mesenchymal stem cells (mBMSCs), and the increased protein was mainly concentrated in the cell nucleus (Fig. 1B). Furthermore, RT-qPCR showed a sustained Foxq1 expression increase in the mBMSCs after osteogenic induction treatment (Fig. 1C).
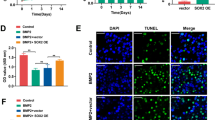
FOXQ1 was upregulated in osteogenesis. A Immunohistology assay results showing FOXQ1 expression in mouse alveolar bone at E15.5 (a1, a2), P7 (b1, b2), and P11 (c1, c2), scale bar 200 μm. B Immunofluorescence staining showed FOXQ1 in mouse bone mesenchymal stem cells after osteogenic induction, scale bar 200 μm. C Relative expression of Foxq1 in mouse bone mesenchymal stem cells increased after osteogenic induction. D RT-qPCR results of Foxq1 expression in Lv5 mBMSCs versus Foxq1-over mBMSCs (a) and Lv3 mBMSCs versus Foxq1-sh mBMSCs (b) and Western blot assay of FOXQ1 levels (c) confirmed manipulation of FOXQ1 expression was achieved through lentivirus transfection of mouse bone mesenchymal stem cells (Lv5, mBMSCs with empty pGLV5 vector control; Lv3, mBMSCs with empty pGLV3 vector control; Foxq1 ↑, mBMSCs with Foxq1 overexpressing vector; Foxq1 ↓, mBMSCs with Foxq1 shRNA expressing vector). *P < 0.05, **P < 0.01, ***P < 0.001
To further study the role of FOXQ1 in the osteogenic differentiation of bone mesenchymal stem cells, FOXQ1 expression was manipulated with a lentivirus plasmid. The RT-qPCR results showed that the Foxq1 mRNA level in the Foxq1-over mBMSC group was 28-fold greater than that in the lv5 mBMSC group (Fig. 1D (a)), while that of the Foxq1-sh mBMSC group was one-half that of the lv3 mBMSC groups (Fig. 1D (b)). These results were further validated with western blot analysis (Fig. 1D (c)).
Increased FOXQ1 levels promote osteogenic differentiation of mouse bone mesenchymal stem cells
Seven days after osteogenic induction, ALP activity assay showed that FOXQ1 expression level was associated with ALP level in mBMSCs (Fig. 2A (f)). After 14 days of osteogenic induction, ARS showed that with increased FOXQ1 levels, mineral deposition was more prominent (Fig. 2A (a, b)), and a decrease in FOXQ1 also inhibited mineral deposition (Fig. 2A (c–d)). The quantification analysis further supported this finding. Compared to the control group, the relative amount of ARS in the Foxq1-overexpressing mBMSCs was 116%, and it was 71% in the Foxq1-sh mBMSCs (Fig. 2A (e)).
Relationship between the expression level of FOXQ1 and the osteogenic differentiation of mouse bone mesenchymal stem cells. A Alizarin red staining of mouse bone mesenchymal stem cells treated with osteogenic induction medium for 14 days (microscopic views: a1–d1, scale bar 200 μm; gross views: a2–d2) and the relative amount of staining (e) showed FOXQ1 expression level influence osteogenic differentiation of mouse bone mesenchymal stem cells. ALP activity assay (f) of mouse bone mesenchymal stem cells treated with osteogenic induction medium for 7 days showed a similar result. B Relative expression of Alp, Ocn, Opg, and Runx2 in mouse bone mesenchymal stem cells at different levels of FOXQ1, 7 days and 14 days after osteogenic induction. Lv5, mBMSCs with empty pGLV5 vector control; Lv3, mBMSCs with empty pGLV3 vector control; Foxq1 ↑, mBMSCs with Foxq1 overexpressing vector; Foxq1 ↓, mBMSCs with Foxq1 shRNA expressing vector. *P < 0.05, **P < 0.01, ***P < 0.001
We then investigated whether FOXQ1 is able to influence the expression of osteogenesis-related molecules with RT-qPCR. On the 7th day and 14th day after osteogenic induction, various osteogenic markers, namely, Alp (Fig. 2B (a1)), Ocn (Fig. 2B (b1)), Opg (Fig. 2B (c1)), and Runx2 (Fig. 2B (d1)), were upregulated in the Foxq1-overexpressing mBMSC group and downregulated in the Foxq1-sh mBMSC group, compared to the levels of the control groups. These results demonstrated a dose-dependent relationship between FOXQ1 levels and osteogenic marker expression in response to the osteogenic induction.
FOXQ1 promotes osteogenic differentiation via Wnt/β-catenin signaling
Wnt/β-catenin is an important pathway in the regulation of osteogenic differentiation. The TOPFlash/FOPFlash assay of β-catenin/TCF/LEF transcriptional activity demonstrated a 5.7-fold increase in the Foxq1-over mBMSCs (Fig. 3A (a)). For the Foxq1-sh mBMSCs, the transcriptional activity was one-half that of the lv3 mBMSCs (Fig. 3A (b)). The subcellular fractional western blot analysis showed that, while the levels of cytoplasmic β-catenin (CTNNB1) protein were similar across the groups (Fig. 3B), a clear dose-dependent relationship was established between FOXQ1 and intranuclear CTNNB1 (Fig. 3B (a, c)). Western blot analysis comparing total CTNNB1 across the groups (Fig. 3B (b, c)) also showed a dose-dependent relationship with FOXQ1, but less prominent than that of intranuclear CTNNB1. These results were further supported by an immunofluorescence study showing an increase in the overlap between DAPI and CTNNB1 in Foxq1-overexpressing mBMSCs, demonstrating an increase in the nuclear translocation of CTNNB1 (Fig. 3D).
FOXQ1 promoted Wnt/β-catenin signaling. a TOP/FOP ratio of mouse bone mesenchymal stem cells expressing different levels of FOXQ1. b Western blot of nuclear and cytoplasmic beta-catenin (CTNNB1) levels in cells with FOXQ1 overexpressed and inhibited (a, C: cytoplasmic, N: nuclear, THOC1 serve as the loading control for nuclear protein, EEF1A1 serve as the loading control for both nuclear and cytoplasmic protein). Total CTNNB1 levels were also compared among groups of mouse bone mesenchymal stem cells (b); relative level of CTNNB1 normalized to loading control was evaluated for total, cytoplasmic, and nuclear CTNNB1(c). c RT-qPCR of Wnt1 (a), Wnt10a (b), and Wnt3 (c) on FOXQ1 expression. d CTNNB1 transnucleation by immunofluorescence. Scale bar for a1, b1, c1, a3, b3, c3, 200 μm; scale bar for a2, b2, c2, a4, b4, c4, 100 μm. Lv5, mBMSCs with empty pGLV5 vector control; Lv3, mBMSCs with empty pGLV3 vector control; Foxq1 ↑, mBMSCs with Foxq1 overexpressing vector; Foxq1 ↓, mBMSCs with Foxq1 shRNA expressing vector
Wnt1, Wnt3, and Wnt10a are indicators of Wnt/β-catenin activity level, and their expression levels were evaluated with RT-qPCR. The overexpression of FOXQ1 upregulated Wnt1 (Fig. 3C (b)), Wnt3 (Fig. 3C (c)), and Wnt10 (Fig. 3C (d)) expression, while FOXQ1 suppression led to the inhibition of their expression (Fig. 3C (b, c, d)).
The treatment with DKK-1 attenuated the FOXQ1-induced increase in osteogenic differentiation (Fig. 4B (a, b)) and osteogenic marker expression (Fig. 4A (a–d)), while 6BIO alleviated the suppression of the osteogenic differentiation (Fig. 4B (c, d)) and expression of the related markers (Fig. 4A (a–d)).
FOXQ1 promoted Wnt/β-catenin signaling by interacting with ANXA2. a RT-qPCR of Alp (a), Ocn (b), Opg (c), and Runx2 (d) showed altered Wnt/β-catenin signaling pathway activity with DKK1, and 6BIO reversed the effect of FOXQ1 overexpression and inhibition. b Alizarin red staining of mouse bone mesenchymal stem cells showed altered Wnt/β-catenin signaling pathway activity with DKK1, and 6BIO reversed the effect of changed FOXQ1 expression on osteogenic differentiation. Scale bar 200 μm. c Co-Ip/MS identified 199 proteins bound with FOXQ1 in both control and Foxq1-overexpressing mouse bone mesenchymal stem cells. d Protein-protein interaction network (a) constructed by the STRING online database based on 15 matches, and these proteins were ranked by their coverage percentage (b) in the Co-Ip/MS results. e The TOP/FOP ratio of mouse bone mesenchymal stem cells showed that depletion of Anxa2 mRNA reversed the promotion of FOXQ1 on Wnt/β-catenin signaling. F Schematic representation illustrates FOXQ1 binding to ANXA2, which increases the transnucleation of CTNNB1, promoting Wnt/β-catenin signaling and osteogenesis
FOXQ1 promotes Wnt/β-catenin signaling via interaction with ANXA2
The coimmunoprecipitation and LC-MS analysis identified various proteins that were pulled down with FOXQ1. A total of 484 proteins were identified in the samples from the lv5 mBMSC group, while 240 proteins were identified in the samples from the Foxq1-over mBMSC group, and 199 proteins were identified in both types of samples to be binding with FOXQ1(Fig. 4C). With the STRING database, 12 proteins were found to be possibly linked to Wnt/β-catenin (Fig. 4D). Among the candidates with coverage greater than 50% (Fig. 4D (b)), ANXA2 was shown in a previous study to enhance the transnucleation of CTNNB1. The TOPFlash/FOPFlash assay also confirmed that Anxa2 mRNA depletion with siAnxa2 reversed the increase in the Wnt/β-catenin signaling in the Foxq1-overexpressing mBMSCs, suggesting that ANXA2 is an integral part of the mechanism by which FOXQ1 promotes Wnt/β-catenin signaling.
Discussion
FOXQ1 is a member of the forkhead box family, and a recent study showed that various members of this family were involved in the osteogenesis process. FOXO1 was found to be an early molecular regulator during mesenchymal stem cell differentiation into osteoblasts [17]. FOXC1 was also demonstrated to promote osteogenesis by regulating RUNX2 during bone formation [18]. Being a transcription factor, FOXQ1 is involved in a wide variety of processes. It has been identified as an oncogenic factor in various carcinomas [19,20,21,22], and it also participated in various physiological processes [8, 12, 13], but its role in the osteogenic differentiation of mesenchymal stem cells is still unknown. In the current study, evident expression of FOXQ1 in bone tissue and mouse bone mesenchymal stem cells was demonstrated. Additionally, osteogenic induction treatment promoted FOXQ1 expression. On the other hand, FOXQ1 upregulation promoted osteogenic differentiation of mouse bone mesenchymal stem cells, while FOXQ1 suppression led to the opposite effects. These results showed that FOXQ1 is also a factor regulating the osteogenesis process.
Our results showed that FOXQ1 overexpression or suppression led to enhanced or attenuated Wnt signaling activity in mBMSCs, respectively. Wnt/β-catenin signaling is an important regulator of osteogenic differentiation [5, 23]. Beta-catenin is the central molecule in this signaling pathway; when intranuclear translocated, it would bound with TCF/LEF and promote the transcription activity of various osteogenesis associated factors [24], including OPG [25], OCN, ALP [26], and RUNX2 [27], promoting osteogenic differentiation of mesenchymal stem cells [26]. We also showed that the influence of FOXQ1 on the osteogenic differentiation of mBMSCs was partially reversed by counterregulating the activity of Wnt/β-catenin signaling, through the Wnt/β-catenin signaling suppressor DKK1 in the Foxq1-over mBMSC group or the Wnt/β-catenin signaling activator 6BIO in the Foxq1-sh mBMSC group. From these results, it can be inferred that FOXQ1 promotes the osteogenic differentiation of mesenchymal stem cells via the Wnt/β-catenin signaling pathway.
Previous studies have shown that FOXQ1 expression is mediated by Wnt/β-catenin [10]. Interestingly, FOXQ1 silencing prevents the nuclear translocation of β-catenin, reducing Wnt/β-catenin signaling [4]. But the detailed mechanism for crosstalk between them is still unclear. We try to explore the possible mechanism with co-IP LC-MS study, and ANXA2 was found to bind with FOXQ1. ANXA2 is a member of the family of calcium-dependent proteins [28] that participate in angiogenesis, ion channel activation, and intercellular interactions [29, 30]. It was also shown to be associated with increased levels of CTNNB1 [31, 32]. A previous study [33] showed that lncRNA–MUF binds ANXA2, which enhances its binding to glycogen synthase kinase 3 beta (GSK3B) and disrupts the formation of the GSK3B/CTNNB1 complex. In our study, Anxa2 mRNA depletion reversed the promoting effect of FOXQ1 overexpression, demonstrating that binding with ANXA2 is important for FOXQ1 function in Wnt signaling regulation to promote osteogenic differentiation of mesenchymal stem cells.
Conclusion
In summary, our study highlights the importance of FOXQ1 as a mediator of mesenchymal stem cell osteogenic differentiation. To the best of our knowledge, this is the first study to demonstrate that FOXQ1 regulates the activities of Wnt/beta-catenin signaling by binding with ANXA2. These results provide novel insights into the mechanism underlying the osteogenic differentiation of mesenchymal stem cells.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- Alp:
-
Alkaline phosphatase
- ANXA2:
-
Annexin A2
- ARS:
-
Alizarin red staining
- Co-Ip:
-
Coimmunoprecipitation
- EEF1A1:
-
Eukaryotic translation elongation factor 1 alpha 1
- FOXQ1:
-
Forkhead box Q1
- Gapdh:
-
Glyceraldehyde-3-phosphate dehydrogenase
- KM mice:
-
Chinese Kunming mice
- LC-MS:
-
Liquid chromatography–mass spectrometry
- mBMSCs:
-
Mouse bone mesenchymal stem cells
- Ocn:
-
Osteocalcin
- Opg:
-
Osteoprotegerin
- Runx2:
-
Runt-related transcription factor 2
- TCF/LEF:
-
T cell factor/lymphoid enhancer factor
- THOC1:
-
Nuclear matrix protein P84
References
Lee SH, Park Y, Song M, Srikanth S, Kim S, Kang MK, et al. Orai1 mediates osteogenic differentiation via BMP signaling pathway in bone marrow mesenchymal stem cells. Biochem Biophys Res Commun. 2016;473:1309–14 Available from: https://pubmed.ncbi.nlm.nih.gov/27086849/. [cited 2020 Jun 26].
Yu J, Xu L, Li K, Xie N, Xi Y, Wang Y, et al. Zinc-modified calcium silicate coatings promote osteogenic differentiation through TGF-β/Smad pathway and osseointegration. Sci Rep. 2017;7 Available from: https://pubmed.ncbi.nlm.nih.gov/28611362/. [cited 2020 Jun 26].
Huang W, Zheng X, Yang X, Fan S. Stimulation of osteogenic differentiation by Saikosaponin-A in bone marrow stromal cells via WNT/β-catenin pathway. Calcif Tissue Int. 2017;100:392–401 Available from: https://pubmed.ncbi.nlm.nih.gov/28185033/. [cited 2020 Jun 26].
Gao Y, Huang E, Zhang H, Wang J, Wu N, Chen X, et al. Crosstalk between Wnt/β-catenin and estrogen receptor signaling synergistically promotes osteogenic differentiation of mesenchymal progenitor cells. PLoS One 2013;8. Available from: https://pubmed.ncbi.nlm.nih.gov/24340027/. [cited 2020 Jun 26].
Glass DA, Karsenty G. In vivo analysis of Wnt signaling in bone. Endocrinology. 2007;148:2630–4 Available from: http://www.ncbi.nlm.nih.gov/pubmed/17395705. [cited 2020 Jun 26].
MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009:9–26 Available from: https://pubmed.ncbi.nlm.nih.gov/19619488/. [cited 2020 Jun 26].
Voronkov A, Krauss S. Wnt/beta-catenin signaling and small molecule inhibitors. Curr Pharm Des. 2013;19:634–64.
Hong HK, Noveroske JK, Headon DJ, Liu T, Sy MS, Justice MJ, et al. The winged helix/forkhead transcription factor Foxq1 regulates differentiation of hair in satin mice. Genesis. 2001;29:163–71 Available from: https://pubmed.ncbi.nlm.nih.gov/11309849/. [cited 2020 Jun 26].
Li Y, Zhang Y, Yao Z, Li S, Yin Z, Xu M. Forkhead box Q1: a key player in the pathogenesis of tumors (review). Int J Oncol. 2016:51–8 Available from: https://pubmed.ncbi.nlm.nih.gov/27176124/. [cited 2020 Jun 26].
Christensen J, Bentz S, Sengstag T, Shastri VP, Anderle P. FOXQ1, a novel target of the Wnt pathway and a new marker for activation of Wnt signaling in solid tumors. PLoS One. 2013;8 Available from: https://pubmed.ncbi.nlm.nih.gov/23555880/. [cited 2020 Jun 26].
Xiang XJ, Deng J, Liu YW, Wan LY, Feng M, Chen J, et al. MIR-1271 inhibits cell proliferation, invasion and EMT in gastric cancer by targeting FOXQ1. Cell Physiol Biochem. 2015;36:1382–94 Available from: https://pubmed.ncbi.nlm.nih.gov/26159618/. [cited 2020 Jun 26].
Zhang T, Wang P, Liu Y, Zhou J, Shi Z, Cheng K, et al. Overexpression of FOXQ1 enhances anti-senescence and migration effects of human umbilical cord mesenchymal stem cells in vitro and in vivo. Cell Tissue Res. 2018;373:379–93 Available from: https://pubmed.ncbi.nlm.nih.gov/29500491/. [cited 2020 Jun 26].
Tu S, Zheng J, Gao X, Guan C, Cai B, Xiang L. The role of Foxq1 in proliferation of human dental pulp stem cell. Biochem Biophys Res Commun. 2018;497:543–9 Available from: https://pubmed.ncbi.nlm.nih.gov/29453987/. [cited 2020 Jun 26].
Wan Y, Zhuo N, Li Y, Zhao W, Jiang D. Autophagy promotes osteogenic differentiation of human bone marrow mesenchymal stem cell derived from osteoporotic vertebrae. Biochem Biophys Res Commun. 2017;488:46–52 Available from: https://pubmed.ncbi.nlm.nih.gov/28476617/. [cited 2020 Jun 26].
Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:1–26 Available from: https://link.springer.com/articles/10.1186/s12859-017-1934-z. [cited 2020 Aug 9].
STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets - PubMed. Available from: https://pubmed.ncbi.nlm.nih.gov/30476243/. [cited 2020 Jun 26].
Teixeira CC, Liu Y, Thant LM, Pang J, Palmer G, Alikhani M. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J Biol Chem. 2010;285:31055–65.
Ouyang N, Li H, Wang M, Shen H, Si J, Shen G. The transcription factor Foxc1 promotes osteogenesis by directly regulating Runx2 in response of intermittent parathyroid hormone (1–34) treatment. Front Pharmacol. 2020;11:1 Available from: https://www.frontiersin.org/article/10.3389/fphar.2020.00592/full. [cited 2020 Aug 11].
Wang W, He S, Ji J, Huang J, Zhang S, Zhang Y. The prognostic significance of FOXQ1 oncogene overexpression in human hepatocellular carcinoma. Pathol Res Pract. 2013;209:353–8 Available from: https://pubmed.ncbi.nlm.nih.gov/23623360/. [cited 2020 Jun 26].
Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K, Kudo K, et al. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70:2053–63 Available from: https://pubmed.ncbi.nlm.nih.gov/20145154/. [cited 2020 Jun 26].
Zhang H, Meng F, Liu G, Zhang B, Zhu J, Wu F, et al. Forkhead transcription factor Foxq1 promotes epithelial-mesenchymal transition and breast cancer metastasis. Cancer Res. 2011;71:1292–301 Available from: https://pubmed.ncbi.nlm.nih.gov/21285253/. [cited 2020 Jun 26].
Fan DM, Feng XS, Qi PW, Chen YW. Forkhead factor FOXQ1 promotes TGF-β1 expression and induces epithelial–mesenchymal transition. Mol Cell Biochem. 2014;397:179–86 Available from: https://pubmed.ncbi.nlm.nih.gov/25287361/. [cited 2020 Jun 26].
Teufel S, Hartmann C. Wnt-signaling in skeletal development. Curr Top Dev Biol. 2019:235–79 Available from: https://pubmed.ncbi.nlm.nih.gov/30902254/. [cited 2020 Jun 26].
Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4:a007906.
Boyce BF, Xing L, Chen D. Osteoprotegerin, the bone protector, is a surprising target for β-catenin signaling. Cell Metab. 2005;2(6):344–5. https://doi.org/10.1016/j.cmet.2005.11.011.
Li Z, Xu Z, Duan C, Liu W, Sun J, Han B. Role of TCF/LEF transcription factors in bone development and osteogenesis. Int J Med Sci. 2018:1415–22 Available from: /pmc/articles/PMC6158667/?report=abstract. [cited 2020 Aug 11].
Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PVN, Komm BS, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–40 Available from: https://pubmed.ncbi.nlm.nih.gov/16043491/. [cited 2020 Aug 11].
Waisman DM. Annexin II tetramer: structure and function. Mol Cell Biochem. 1995;149–150:301–22 Available from: https://pubmed.ncbi.nlm.nih.gov/8569746/. [cited 2020 Jun 26].
Chiang Y, Rizzino A, Sibenaller ZA, Wold MS, Vishwanatha JK. Specific down-regulation of annexin II expression in human cells interferes with cell proliferation. Mol Cell Biochem. 1999;199:139–47 Available from: https://pubmed.ncbi.nlm.nih.gov/10544962/. [cited 2020 Jun 26].
Liu L, Tao J-Q, Zimmerman U-JP. Annexin II binds to the membrane of A549 cells in a calcium-dependent and calcium-independent manner. Cell Signal. 1997;9:299–304.
Tang T, Guo C, Xia T, Zhang R, Zen K, Pan Y, et al. LncCCAT1 promotes breast cancer stem cell function through activating WNT/β-catenin signaling. Theranostics. 2019;9:7384.
Sarkar S, Swiercz R, Kantara C, Hajjar KA, Singh P. Annexin A2 mediates up-regulation of NF-κB, β-catenin, and stem cell in response to progastrin in mice and HEK-293 cells. Gastroenterology. 2011;140:583–95.
Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y, et al. Mesenchymal stem cells promote hepatocarcinogenesis via lncRNA–MUF interaction with ANXA2 and miR-34a. Cancer Res. 2017;77:6704–16.
Acknowledgements
Not applicable
Funding
The present study was supported by grants from the National Natural Science Foundation of China (81800961), Science and Technology Program of Guangdong Province (2016B030229003), Natural Science Foundation of Guangdong Province (2018A030313098), and Medical Scientific Research Foundation of Guangdong Province (B2020158).
Author information
Authors and Affiliations
Contributions
Lusai Xiang and Junming Zheng have made substantial contributions to the conception and design of the work. Bin Cai has made substantial contribution to the design of the work. Tingting Ai and Mengdan Zhang contributed to the acquisition analysis and interpretation of data. All authors have approved the submitted version of the article and have agreed to be personally accountable for the author’s own contributions and to ensure that questions related to the accuracy or integrity of any part of the work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the Hospital of Stomatology, Sun Yat-sen University (ERC-2013-15; Guangzhou, China).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Xiang, L., Zheng, J., Zhang, M. et al. FOXQ1 promotes the osteogenic differentiation of bone mesenchymal stem cells via Wnt/β-catenin signaling by binding with ANXA2. Stem Cell Res Ther 11, 403 (2020). https://doi.org/10.1186/s13287-020-01928-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-020-01928-9