Abstract
Background
MicroRNAs (miRNAs) play a key role in regulating cell differentiation. In the present study, we aimed to explore the role of miR-140-5p in odontoblastic differentiation of dental pulp stem cells (DPSCs).
Methods
DPSCs from normal human impacted third molars were isolated and cultured. After overexpression or silencing of miR-140-5p in DPSCs, activity, proliferation, and odontoblastic differentiation of DPSCs were evaluated. The possible target gene of miR-140-5p was verified by luciferase reporter gene assay. Using gene transfection technology, RT-CPR, and Western blot to confirm miR-140-5p regulates the odontoblastic differentiation of DPSCs through Wnt1/β-catenin signaling.
Results
We found the expression of miR-140-5p decreased in the differentiated DPSCs for odontoblastic cells, and at the same time, the expressions of Wnt1 and β-catenin increased. Wnt1 was the target gene of miR-140-5p which was confirmed by luciferase reporter gene system. miR-140-5p overexpression suppressed the expression of Wnt1. miR-140-5p inhibitor could promote the odontoblastic differentiation of DPSCs. miR-140-5p mimic could weaken the odontoblastic differentiation of DPSCs, which could be reversed by the overexpression of Wnt1.
Conclusion
Our data demonstrated that miR-140-5p regulates the odontoblastic differentiation of DPSCs via targeting Wnt1/β-catenin signaling. Therefore, miR-140-5p might be a molecular target to regulate the odontoblastic differentiation for the therapeutic agents in dental medicine.
Similar content being viewed by others
Background
The exposure of the pulp can occur as a consequence of deep caries, traumatic injuries, or iatrogenic injury and often results in inflammation or death of the pulp [1]. Endodontically treated teeth become devitalized, brittle, and susceptible to postoperative fracture or other complications, including re-infections [2]. Therefore, new strategies are needed in order to try to regenerate the original tissue and subsequently restore the dental pulp properties [1]. Dental pulp stem cells (DPSCs), as a type of fibroblast, are present in the pulp. It has multidirectional differentiation potential and is responsible for the self-repairing function of the dental pulp tissue. DPSCs have become a hot spot in stomatological research in the world [3]. Studies have shown that DPSCs can differentiate into osteoblasts, odontoblasts, adipocytes, muscle cells, and nerve cells, with strong proliferative capacity and high differentiation potential [4]. The regulation mechanism of DPSC-directed differentiation has been a hot topic in the field, and there are still many difficulties to be solved. This also greatly limits the clinical application of DPSCs.
MicroRNAs are one of the most abundant gene-regulating molecules that may affect the export of many protein-coding genes in multicellular organisms. There is increasing evidence that microRNAs are involved in a wide range of underlying cellular processes such as cell differentiation, proliferation, growth, migration, and apoptosis as well as carcinogenesis or inhibition of cancers [5,6,7,8]. miR-140-5p is a member of the microRNAs family. Recent studies have shown that miR-140-5p is involved in the proliferation and differentiation of cells [9,10,11,12]. In the preliminary experiments of inducing differentiation of DPSCs into odontoblasts, we observed the expression of miR-140-5p was significantly decreased in the induced differentiation group. The target gene of miR-140-5p was analyzed and predicted using the online software targetscan (http://www.targetscan.org). The results showed that Wnt1 may be one of the target genes of miR-140-5p (more detail is shown in Additional file 1). Our previous study found β-catenin was also involved in the odontoblast differentiation of DPSCs [6]. The Wnt/β-catenin signaling pathway is widely distributed in invertebrates and vertebrates and is a highly conserved signaling pathway during species evolution [13]. Wnt/β-catenin signaling plays a crucial role in the early development of embryos, organ formation, tissue regeneration, and other physiological processes [14,15,16,17,18,19]. In summary, this study proposes that miR-140-5p regulates the odontoblastic differentiation of DPSCs via targeting Wnt1/β-catenin signaling. In this study, DPSCs from normal human impacted third molars were isolated and cultured. After overexpression or silencing of miR-140-5p in DPSCs, MTT assay and Ki67 immunofluorescence were used to evaluate the effect of miR-140-5p on the activity and proliferation of DPSCs. The possible target gene of miR-140-5p was verified by luciferase reporter gene assay. Using gene transfection technology, RT-CPR, and Western blot to confirm miR-140-5p regulates the odontoblastic differentiation of DPSCs through Wnt1/β-catenin signaling.
Materials and methods
Cell culture
The cell culture process was performed as described in our previous studies [6, 20] and approved by the Ethics Committee of the Affiliated Hospital of Nantong University. Briefly, normal human impacted third molars were collected from patients of 14~22 years of age (n = 10) after they had given informed consent. The pulp was digested in a solution of 3 mg/mL collagenase type I for 1 h at 37 °C. Single-cell suspensions of dental pulp were seeded into 25-cm2 culture dishes and cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C under 5% CO2. Cells were passaged at the ratio of 1:3 when they reached 85–90% confluence. The cell populations were characterized by positive staining with anti-CD34, STRO-1, and c-kit, and by the absence of CD45. Cells from the fourth passage were used in all experiments.
Odontoblastic differentiation
The odontoblastic differentiation process was performed as described in our previous study [6]. Briefly, DPSCs from the fourth passages were cultured in odontogenic differentiation medium containing a Minimum Essential Medium (Invitrogen, Carlsbad, CA), 15% FBS, 10 mmol/L β-glycerophosphate, 50 mg/mL α-ascorbic acid, 10 nmol/L dexamethasone (Sigma-Aldrich, St. Louis, MO), 0.292 mg/mL glutamine, 100 μg/mL streptomycin, and 100 U/mL penicillin for 14 days.
The expression of miR-140-5p was detected by qRT-PCR
The RNA in cells was extracted using TaqMan miRNA isolation kit, then the expression of mature miR-140-5p was detected by the TaqMan miRNA assay and TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA). U6 was used as the internal reference. The qRT-PCR relative quantitative method was used to analyze the experimental result.
Cell transfection
The miR-140-5p mimic (abm, Canada), miR-140-5p inhibitor (abm, Canada), corresponding negative control, restructuring plasmid pcDNA3.1-Wnt1, and empty vector control pcDNA3.1 (GenePharma, Shanghai, China) were transfected into DPSCs using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction.
Alizarin red S staining
The staining process was performed as described in our previous study [21]. The differentiated cells were fixed with 4% paraformaldehyde (PFA) for 30 min and washed with PBS. Subsequently, cells were incubated with 2% Alizarin red S solution for 10 min at room temperature under gentle agitation. Cells were then washed with deionized water to remove excess staining. Mineralization was quantified by extracting the Alizarin red S stain with 100 mM cetylpyridinium chloride solution (Sigma-Aldrich) at room temperature. The absorbance of the extracted Alizarin red S stain was measured at 570 nm.
Cell activity assay
DPSC activity was measured by using methyl-thiazole-tetrazolium (MTT) assay as described by Peng et al. [22]. MTT (AMRESCO) solution (5 mg/mL) was added to the cultures (10 μL MTT solution per 100 μL medium), followed by the incubation with 5% CO2 at 37 °C for 4 h. A solution of sodium dodecyl sulfate (SDS) was added to the cultures, which was incubated for 20 h at 37 °C in 5% CO2. The optical density (OD) was read on a Universal Microplate Reader (Synergy 2, Bio-Tek Instruments, Inc., USA) using a test wavelength of 570 nm.
Cell proliferation assay
DPSC proliferation was measured by using Ki67 immunofluorescence. Cells were first fixed in 4% PFA, followed by incubation in hydrogen peroxide, and then incubated with the rat anti-Ki67 antibody (1:800, Abcam, Cambridge, UK) overnight. The following day, the cells were labeled the secondary antibody with Alexa Fluor 568-conjugated goat anti-rat (1:1000, Molecular Probes). The cell nuclei were counter-stained with Hoechst33342 for 30 min. Positive cells were observed under a fluorescent microscope.
Western blot analysis
The cell total protein analysis process was performed as described in our previous study [20]. Briefly, the separated proteins were transferred onto polyvinylidene difluoride membranes, then the membranes were blocked with 5% nonfat milk and incubated with primary antibodies and second antibodies in turn. The following primary antibodies were used: rabbit anti-Wnt1(1:1000, Abcam), rabbit anti-β-catenin (1:1000, Abcam), rabbit anti-dentin sialophosphoprotein (DSPP) (1:1000, Abcam), rabbit anti-dentin matrix protein-1 (DMP-1) (1:1000, Abcam), and mouse anti-β-actin (1:1000, Santa Cruz). The second antibodies were goat-anti-rabbit or goat-anti-mouse horseradish peroxidase-conjugated IgG (1:1500, Abcam).
A Nuclear Protein Extraction Kit (Active Motif, Carlsbad, CA, USA) was used to extract the nuclear fractions according to the manufacturer’s instructions. The PVDF membranes were probed with specific antibodies: rabbit anti-β-catenin (1:1000, Abcam) and mouse anti-Lamin B1 (1:2000, Abcam) overnight at 4 °C. Then, they are followed by incubation with the second antibodies: goat-anti-rabbit or goat-anti-mouse horseradish peroxidase-conjugated IgG (1:1500, Abcam).
The target gene of miR-140-5p was detected by luciferase reporter gene system
Online software TargetScan (http://www.targetscan.org) was used to predict the target gene of miR-140-5p, and the results showed Wnt1 might be the target gene. The wild-type (WT) 3′-UTR of Wnt1 mRNA has a putative miR-140-5p-binding site. The mutant-type (MuT) 3′-UTR of Wnt1 was constructed. The WT 3′-UTR of Wnt1 and MuT 3′-UTR of Wnt1 were inserted into pMIRGLO vectors (Promega Corporation, Madison, WI, USA). The miR-140-5p mimic, miR-negative control, WT 3′-UTR of Wnt1 vector, and MuT 3′-UTR of Wnt1 vector were co-transfected into the HEK293 cells. Following cell transfection for 48 h, luciferase activity was detected using a dual-luciferase reporter assay system (E1910; Promega Corporation).
Statistical analysis
All the data were expressed by mean ± SD in this study and analyzed using statistical software SPSS 21.0. The difference among the groups was estimated by one-way ANOVA and compared using the LSD method. P < 0.05 was considered statistically significant.
Results
The expression of miR-140-5p is significantly decreased in the induced differentiation group
After differentiation of day 1, day 3, day 7, and day 14, the expressions of miR-140-5p in the differentiation groups were lower than that in the control group, and the difference was statistically significant (P < 0.05). But there were no significant differences among day 1, day 3, day 7, and day 14 (P > 0.05) (Fig. 1).
The expression of miR-140-5p was detected by qRT-PCR. After differentiation of day 1, day 3, day 7, and day 14, the expressions of miR-140-5p in the differentiation groups were lower than that in the control group. But there were no significant differences among day 1, day 3, day 7, and day 14. *vs. the control group, P < 0.05. n = 6
miR-140-5p has no effects on cell activity and proliferation
Following cell transfection for 48 h, the expression of miR-140-5p was detected by qRT-PCR; the cell activity and proliferation were measured by MMT and Ki67 immunofluorescence, respectively. Compared with the control, mimic negative control (NC), and inhibitor NC groups, the expressions of miR-140-5p were increased in the mimic group and decreased in the inhibitor group, and the differences were statistically significant (P < 0.05) (Fig. 2a). There were no significant differences in OD values of the control, mimic NC, inhibitor NC, mimic, and inhibitor groups (P > 0.05) (Fig. 2b). There were also no significant differences in Ki67+ cell number in the five groups (P > 0.05) (Fig. 2c).
The effects of miR-140-5p on cell activity and proliferation. a The expression of miR-140-5p was detected by qRT-PCR. Compared with the control, mimic NC, and inhibitor NC groups, the expression of miR-140-5p increased in the mimic group and decreased in the inhibitor group. b The cell activity was measured by MTT. OD values of the control, mimic, mimic NC, inhibitor, and inhibitor NC groups had no differences. c The cell proliferation was measured by Ki67 immunofluorescence. There were no significant differences in Ki67+ cell number of the control, mimic, mimic NC, inhibitor, and inhibitor NC groups. *vs. the control group, P < 0.05; #vs. the mimic NC group, P < 0.05; @vs. the inhibitor NC group, P < 0.05; &vs. the mimic group, P < 0.05. n = 6
miR-140-5p inhibit the odontoblastic differentiation of DPSCs
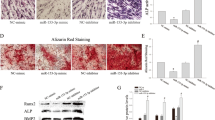
Following cell transfection for 48 h, the cells were induced to odontoblastic differentiation for 14 days. Alizarin red S staining showed the mineralized matrix deposition was the most in the inhibitor group and the least in the mimic group (P < 0.05); the differences among the control, mimic NC, and inhibitor NC groups were not statistically significant (P > 0.05) (Fig. 3a). DSPP and DMP-1 proteins, marker of odontoblast, were detected by Western blot (see Additional file 2). The results showed the expressions of DSPP and DMP-1 proteins were the highest in the inhibitor group and the lowest in the mimic group (P < 0.05); the differences among the control, mimic NC, and inhibitor NC groups were not statistically significant (P > 0.05) (Fig. 3b).
miR-140-5p inhibited the odontoblastic differentiation of DPSCs. a Alizarin red S staining showed the mineralized matrix deposition was the most in the inhibitor group and the least in the mimic group; the differences among the control, mimic NC, and inhibitor NC groups were not statistically significant. b The DMP-1 and DSPP proteins were detected by Western blot. The results showed the expressions of DSPP and DMP-1 proteins were the highest in the inhibitor group and the lowest in the mimic group; the differences among the control, mimic NC, and inhibitor NC groups were not statistically significant. *vs. the control group, P < 0.05; #vs. the mimic NC group, P < 0.05; @vs. the inhibitor NC group, P < 0.05; &vs. the mimic group, P < 0.05. Bar = 50 μm. n = 6
Wnt1 was the target gene of miR-140-5p
Luciferase reporter gene detection results suggested that the luciferase activity of the mimic group was significantly lower than that of the mimic NC group (P < 0.05) (Fig. 4a). Western blot analysis showed the expressions of Wnt1 and total or nuclear β-catenin proteins were the highest in the inhibitor group and the lowest in the mimic group (P < 0.05); the differences among the control, mimic NC, and inhibitor NC groups were not statistically significant (P > 0.05) (Fig. 4b, c).
Wnt1 was the target gene of miR-140-5p. a Luciferase reporter gene detection results suggested that the luciferase activity of the mimic group was significantly lower than that of the mimic NC group. b Western blot analysis showed the expressions of Wnt1 and total β-catenin proteins were the highest in the inhibitor group and the lowest in the mimic group; the differences among the control, mimic NC, and inhibitor NC groups were not statistically significant. c Western blot analysis showed the expression of nuclear β-catenin protein was the highest in the inhibitor group and the lowest in the mimic group; the differences among the control, mimic NC, and inhibitor NC groups were not statistically significant. *vs. the control group, P < 0.05; #vs. the mimic NC group, P < 0.05; @vs. the inhibitor NC group, P < 0.05; &vs. the mimic group, P < 0.05. n = 6
Overexpression of Wnt1 reverses the inhibition of odontoblastic differentiation induced by miR-140-5p
Following cell transfection for 48 h, the cells were induced to odontoblastic differentiation for 14 days. Alizarin red S staining showed the mineralized matrix deposition was the least in the mimic group and the most in the pcDNA3.1-Wnt1 group (P < 0.05). The number of positive cells in the mimic+pcDNA3.1-Wnt1 group was similar to that of the control, mimic NC, and pcDNA3.1 NC groups; the differences among them were not statistically significant (P > 0.05) (Fig. 5a). DSPP and DMP-1 are necessary for the proper biomineralization of cementum, dentin, or enamel, which are usually used as the markers of odontoblastic differentiation. Western blot analysis showed the expressions of DMP-1 and DSPP proteins were the highest in the pcDNA3.1-Wnt1 group and the lowest in the mimic group (P < 0.05). The expressions of DMP-1 and DSPP proteins in the mimic+pcDNA3.1-Wnt1 group were similar to those of the control, mimic NC, and pcDNA3.1 NC groups; the differences among them were not statistically significant (P > 0.05) (Fig. 5b).
Overexpression of Wnt1 reversed the inhibition of odontoblastic differentiation induced by miR-140-5p. a Alizarin red S staining showed the mineralized matrix deposition was the least in the mimic group and the most in the pcDNA3.1-Wnt1 group. The percent of positive cells in the pcDNA3.1-Wnt1+mimic group was similar to that of the control, mimic NC, and pcDNA3.1 NC groups; the differences among them were not statistically significant. b Western blot analysis showed the expressions of DSPP and DMP-1 proteins were the highest in the pcDNA3.1-Wnt1 group and the lowest in the mimic group. The expressions of DSPP and DMP-1 proteins in the pcDNA3.1-Wnt1+mimic group were similar to those of the control, mimic NC, and pcDNA3.1 NC groups; the differences among them were not statistically significant. *vs. the control group, P < 0.05; #vs. the mimic NC group, P < 0.05; @vs. the pcDNA3.1 NC group, P < 0.05; &vs. the mimic group, P < 0.05; $vs. the mimic+pcDNA3.1-Wnt1 group, P < 0.05. n = 6
Discussion
In this study, first, we found the expression of miR-140-5p was significantly decreased in the induced odontoblast differentiation. Based on this result, we downregulated or overexpressed miR-140-5p in DPSCs and found miR-140-5p knockdown upregulated odontoblast differentiation, while miR-140-5p overexpression had a negative effect on odontoblast differentiation in DPSCs. Furthermore, Wnt1, the target gene of miR-140-5p, was confirmed. We approved that miR-140-5p regulated the odontoblast differentiation of DPSCs through the Wnt/β-catenin pathway.
DPSCs can be simply isolated from freshly extracted teeth. These cells can be highly proliferated and multilineage-differentiated. Moreover, their clinical utility is attributable to their simplicity and convenience of isolation, lack of ethical controversy, and low immunogenicity and is used as autologous transplants for dentin regeneration and periodontal tissue regeneration [20]. However, how to regulate the directed differentiation of DPSCs is still a major problem for researchers. miR-140-5p is a member of the microRNAs family. Recent studies have shown that miR-140-5p is involved in the proliferation and differentiation of cells [9,10,11,12]. In the study of osteogenic or adipogenic differentiation of human adipose-derived mesenchymal stem cells, it was found that low expression of miR-140-5p could promote osteogenic differentiation [23]. Hwang et al. reported that miR-140-5p suppressed BMP2-mediated osteogenesis in undifferentiated human mesenchymal stem cells [11]. Sun et al. found miR-140-5p-mediated regulation of the proliferation and differentiation in DPSCs occurred through the lipopolysaccharide/Toll-like receptor 4 signaling pathway. Their results showed overexpression of miR-140-5p enhanced proliferation of DPSCs and inhibited DPSC differentiation, whereas suppression of miR-140-5p produced the opposite effects [12]. Our results indicated that low expression of miR-140-5p could promote odontoblast differentiation of DPSCs, whereas overexpression of miR-140-5p inhibited odontoblast differentiation, but the expression of miR-140-5p had no effect on the activity and proliferation of DPSCs, which was different from Sun et al. [12]; this difference may be due to the different detection condition. All the results indicated that miR-140-5p was a key miRNA that regulated DPSC proliferation and differentiation.
Furthermore, Wnt1, the target gene of miR-140-5p was confirmed in our study by luciferase reporter gene detection system. Western blot analysis showed the overexpression of miR-140-5p inhibited the expressions of Wnt1 and β-catenin, whereas the suppression of miR-140-5p produced the opposite effects. The overexpression of Wnt1 could reverse the inhibition of odontoblastic differentiation induced by miR-140-5p. The classical Wnt/β-catenin signaling pathway plays a crucial role in the early development of embryos, organ formation, tissue regeneration, and other physiological processes [14,15,16,17,18,19]. Meanwhile, more and more researches support that the Wnt/β-catenin signaling pathway regulates stem cell self-renewal, proliferation, and differentiation [24,25,26] and was involved in dental tissue development [27,28,29]. Our previous study found β-catenin was involved in the odontoblast differentiation of DPSCs, too [6]. In this study, the total and nuclear β-catenin protein levels were the highest in the inhibitor group and the lowest in the mimic group, which indicated miR-140-5p was involved in β-catenin activation. Combining the results of this study, it was believed that miR-140-5p regulated the odontoblast differentiation of DPSCs via the Wnt1/β-catenin signaling pathway. Since the expression level of miR-140-5p is naturally downregulated during odontoblastic differentiation of DPSCs, whether it is still meaningful to deliver inhibitor to the site of pulp regeneration needs further studies to confirm it although the result in this study indicates that miR-140-5p inhibitor promotes the odontoblastic differentiation of DPSCs in vitro.
Conclusion
In summary, our data demonstrated that miRNA-140-5p regulates the odontoblastic differentiation of DPSCs via targeting the Wnt1/β-catenin signaling pathway. Therefore, miRNA-140-5p might be a molecular target to regulate the odontoblastic differentiation for therapeutic agents in dental medicine.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Coimbra P, Alves P, Valente TA, Santos R, Correia IJ, Ferreira P. Sodium hyaluronate/chitosan polyelectrolyte complex scaffolds for dental pulp regeneration: synthesis and characterization. Int J Biol Macromol. 2011;49(4):573–9.
Sun HH, Jin T, Yu Q, Chen FM. Biological approaches toward dental pulp regeneration by tissue engineering. J Tissue Eng Regen Med. 2011;5(4):e1–16.
Hilkens P, Meschi N, Lambrechts P, Bronckaers A, Lambrichts I. Dental stem cells in pulp regeneration: near future or long road ahead? Stem Cells Dev. 2015;24(14):1610–22.
Mead B, Logan A, Berry M, Leadbeater W, Scheven BA. Concise review: dental pulp stem cells: a novel cell therapy for retinal and central nervous system repair. Stem Cells. 2017;35(1):61–7.
Park MG, Kim JS, Park SY, Lee SA, Kim HJ, Kim CS, et al. MicroRNA-27 promotes the differentiation of odontoblastic cell by targeting APC and activating Wnt/beta-catenin signaling. Gene. 2014;538(2):266–72.
Song Y, Liu X, Feng X, Gu Z, Gu Y, Lian M, et al. NRP1 accelerates odontoblast differentiation of dental pulp stem cells through classical Wnt/beta-catenin signaling. Cell Reprogram. 2017;19(5):324–30.
Adelman K, Egan E. Non-coding RNA: more uses for genomic junk. Nature. 2017;543(7644):183–5.
Ren B. Transcription: enhancers make non-coding RNA. Nature. 2010;465(7295):173–4.
Li H, Guan SB, Lu Y, Wang F. MiR-140-5p inhibits synovial fibroblasts proliferation and inflammatory cytokines secretion through targeting TLR4. Biomed Pharm. 2017;96:208–14.
Zhang Y, Xu J. MiR-140-5p regulates hypoxia-mediated human pulmonary artery smooth muscle cell proliferation, apoptosis and differentiation by targeting Dnmt1 and promoting SOD2 expression. Biochem Biophys Res Commun. 2016;473(1):342–8.
Hwang S, Park SK, Lee HY, Kim SW, Lee JS, Choi EK, et al. miR-140-5p suppresses BMP2-mediated osteogenesis in undifferentiated human mesenchymal stem cells. FEBS Lett. 2014;588(17):2957–63.
Sun DG, Xin BC, Wu D, Zhou L, Wu HB, Gong W, et al. miR-140-5p-mediated regulation of the proliferation and differentiation of human dental pulp stem cells occurs through the lipopolysaccharide/toll-like receptor 4 signaling pathway. Eur J Oral Sci. 2017;125(6):419–25.
DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308(5723):826–33.
Taciak B, Pruszynska I, Kiraga L, Bialasek M, Krol M. Wnt signaling pathway in development and cancer. J Physiol Pharmacol. 2018;69:2.
Yodthong T, Kedjarune-Leggat U, Smythe C, Wititsuwannakul R, Pitakpornpreecha T. l-Quebrachitol Promotes the Proliferation, Differentiation, and Mineralization of MC3T3-E1 Cells: Involvement of the BMP-2/Runx2/MAPK/Wnt/beta-Catenin Signaling Pathway. Molecules. 2018;23(12):E3086.
Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359(6380):1118–23.
Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–99.
Chanda B, Ditadi A, Iscove NN, Keller G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell. 2013;155(1):215–27.
Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–205.
Zhang J, Lu X, Feng G, Gu Z, Sun Y, Bao G, et al. Chitosan scaffolds induce human dental pulp stem cells to neural differentiation: potential roles for spinal cord injury therapy. Cell Tissue Res. 2016;366(1):129–42.
Feng X, Feng G, Xing J, Shen B, Li L, Tan W, et al. TNF-alpha triggers osteogenic differentiation of human dental pulp stem cells via the NF-kappaB signalling pathway. Cell Biol Int. 2013;37(12):1267–75.
Peng YP, Qiu YH, Lu JH, Wang JJ. Interleukin-6 protects cultured cerebellar granule neurons against glutamate-induced neurotoxicity. Neurosci Lett. 2005;374(3):192–6.
Li Z, Jin C, Chen S, Zheng Y, Huang Y, Jia L, et al. Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol Cell Biochem. 2017;433(1–2):51–60.
L’Episcopo F, Tirolo C, Peruzzotti-Jametti L, Serapide MF, Testa N, Caniglia S, et al. Neural stem cell grafts promote astroglia-driven neurorestoration in the aged parkinsonian brain via Wnt/beta-catenin signaling. Stem Cells. 2018;36(8):1179–97.
Yang Z, Balic A, Michon F, Juuri E, Thesleff I. Mesenchymal Wnt/beta-catenin signaling controls epithelial stem cell homeostasis in teeth by inhibiting the antiapoptotic effect of Fgf10. Stem Cells. 2015;33(5):1670–81.
Manton KJ, Leong DF, Cool SM, Nurcombe V. Disruption of heparan and chondroitin sulfate signaling enhances mesenchymal stem cell-derived osteogenic differentiation via bone morphogenetic protein signaling pathways. Stem Cells. 2007;25(11):2845–54.
Yang G, Zhou J, Teng Y, Xie J, Lin J, Guo X, et al. Mesenchymal TGF-beta signaling orchestrates dental epithelial stem cell homeostasis through Wnt signaling. Stem Cells. 2014;32(11):2939–48.
Kim TH, Bae CH, Lee JC, Ko SO, Yang X, Jiang R, et al. β-catenin is required in odontoblasts for tooth root formation. J Dent Res. 2013;92(3):215–21.
Chen J, Lan Y, Baek JA, Gao Y, Jiang R. Wnt/beta-catenin signaling plays an essential role in activation of odontogenic mesenchyme during early tooth development. Dev Biol. 2009;334(1):174–85.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China, Grant/Award Number: 81500809.
Author information
Authors and Affiliations
Contributions
GF and XF conceived and designed the experiments. XL, XC, JX, ML, DH, and YL performed the experiments. XL and XF analyzed the data. XL and XC wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
The outcome of target gene prediction for miR-140-5p. The results showed that Wnt1 may be one of the target genes of miR-140-5p (red part). (XLSX 58 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lu, X., Chen, X., Xing, J. et al. miR-140-5p regulates the odontoblastic differentiation of dental pulp stem cells via the Wnt1/β-catenin signaling pathway. Stem Cell Res Ther 10, 226 (2019). https://doi.org/10.1186/s13287-019-1344-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-019-1344-4