Abstract
Purpose
To investigate the reproducibility of radiomics features extracted from two-dimensional regions of interest (2D ROIs) versus whole lung (3D) ROIs in repeated in-vivo fetal magnetic resonance imaging (MRI) acquisitions.
Methods
Thirty fetal MRI scans including two axial T2-weighted acquisitions of the lungs were analysed. 2D (lung at the level of the carina) and 3D (whole lung) ROIs were manually segmented using ITK-Snap. Ninety-five radiomics features were extracted from 2 and 3D ROIs in initial and repeat acquisitions using Pyradiomics. Radiomics feature intra-class correlation coefficients (ICC) were calculated between 2 and 3D ROIs in the initial acquisition, and between 2 and 3D ROIs in repeated acquisitions, respectively.
Results
MRI data of 11 (36.7%) female and 19 (63.3%) male fetuses acquired at a median 25 + 0 gestational weeks plus days (GW) (interquartile range [IQR] 23 + 4 − 27 + 0 GW) were assessed. Median radiomics feature ICC between 2 and 3D ROIs in the initial MRI acquisition was 0.733 (IQR 0.313–0.814, range 0.018–0.970). ICCs between radiomics features extracted using 3D ROIs in initial and repeat acquisitions (median 0.908 [IQR 0.824–0.929, range 0.335–0.996]) were significantly higher compared to 2D ROIs (0.771 [0.699–0.835, 0.048–0.965]) (p < 0.001).
Conclusion
Fetal MRI radiomics features extracted from 3D whole lung segmentation masks showed significantly higher reproducibility across repeat acquisitions compared to 2D ROIs. Therefore, fetal MRI whole lung radiomics features are robust diagnostic and potentially prognostic tools in the image-based in-vivo quantitative assessment of lung development.
Key points
-
Standardised fetal MRI delivers reproducible quantitative whole lung radiomics features.
-
2D lung ROI radiomics features are not representative of the whole fetal lung.
-
Whole lung radiomics features are better reproducible than 2D ROI features.
-
Whole lung radiomics features are more sensitive to subtle differences between normal and pathological fetal lung development compared to 2D lung ROI features.
-
Fetal MRI whole lung radiomics may improve non-invasive in-vivo lung development assessment.
Similar content being viewed by others
Background
Prenatal detection of pathologic lung development is a prerequisite for timely resource allocation, including delivery in a centre offering advanced neonatal respiratory support techniques such as extra-corporeal membrane oxygenation. Currently, non-invasive in-vivo imaging assessment of fetal lung growth relies primarily on fetal ultrasound or magnetic resonance imaging (MRI) based lung volumetry [1]. This approach is limited by wide gestational age and body volume-adjusted normal lung volume ranges that may only inaccurately identify fetuses at risk for postnatal respiratory insufficiency [2, 3]. Research on fetal MRI signal intensity-based analysis of lung maturity has produced variable results, thus far precluding its clinical translation [4].
The use of radiomics, a quantitative image analysis method that is used to extract a large number of features from medical image data, has been proposed to increase the diagnostic and prognostic imaging yield in a variety of conditions [5,6,7]. Recently, studies have highlighted the potential of radiomics for the identification of fetuses with abnormal lung development and texture-based neonatal respiratory distress prediction [8,9,10]. Thus far, fetal lung radiomics features have been extracted from two-dimensional (2D), representative-appearing regions of interest (ROI) in fetal lung ultrasound images [10]. However, it is unclear if features extracted from 2D ROIs are sufficient to reflect the developmental status of the entire fetal lung. In addition, reproducibility of radiomics features in test–retest conditions is a precondition for their safe and meaningful application according to the Radiomics Quality Score proposed by Lambin et al. [11]. To date, there is a lack of evidence regarding the reproducibility of radiomics features extracted from the fetal lung using 2D ROIs, limiting the wider application of radiomics in fetal lung imaging and beyond. Fetal MRI offers the opportunity to obtain three-dimensional (3D) image data of the developing lungs in a standardised fashion, and may facilitate reproducible radiomics-based quantitative assessment of lung development.
Therefore, this study had two aims: First, to investigate whether fetal MRI lung radiomics features extracted from 2D ROIs are representative of features extracted from whole lung segmentation masks (3D ROIs). Second, to investigate the reproducibility of lung radiomics features extracted from 2 and 3D ROIs in repeated standardised fetal MRI acquisitions.
Methods
This retrospective, single-centre study was approved by the Ethics Committee of the Medical University of Vienna (1232/2022). The requirement to obtain informed consent was waived. A part of the study cohort (n = 29) has been previously reported in a study that did not include 2D radiomics feature analyses [12].
Patients
A retrospective search of the hospital picture and archiving system (PACS) was conducted to identify thirty cases that underwent clinically indicated fetal MRI including two axial T2-weighted acquisitions of the developing lung between January of 2016 and February of 2022. The sample size was chosen in accordance with previous test–retest studies investigating radiomics feature reproducibility [13,14,15]. In order to assess radiomics feature reproducibility, cases with normal and diffuse lung pathology (e.g. pulmonary hypoplasia due to premature rupture of membranes) at different gestational ages were included. Cases were excluded from analysis for lack of ultrasound-based gestational age estimation, presence of a focal lung malformation, incomplete lung representation on fetal MRI, lung tissue visible on five or fewer MRI slices, or presence of (fetal or maternal) motion artifacts.
Fetal MRI
Fetal MRI data from clinically indicated scans were retrospectively identified. Repeated standardised axial T2-weighted acquisitions of the lungs were routinely performed for e.g. lung volumetry or data collection for super-resolution body imaging according to the clinical indication and in accordance with ISUOG guidelines [16]. One 1.5 T MRI scanner (Ingenia, Philips Healthcare, Best, The Netherlands) was used for all exams. Axial T2-weighted acquisitions were acquired in a standardised fashion using a body coil and the following parameters: 200 to 300 mm field of view, 3 to 4 mm slice thickness (thinner slices used in early gestation), 0.3 to 0.4 mm gap, 256 × 256 matrix, shortest (7536.2 to 31,575 ms) repetition time, 100 ms echo time, and 90° flip angle. Specific absorption rates were less than 2W/kg for all cases. Fetal MRI scans were performed without administration of sedation or contrast medium. The time in minutes between initial and repeat axial T2-weighted acquisitions was calculated for each case. Gestational age in weeks plus days (GW) post menstruationem at the time of the fetal MRI scan was calculated based on the first fetal ultrasound examination.
Radiomics
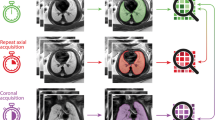
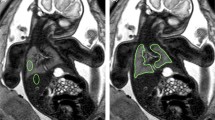
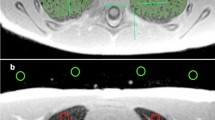
Anonymised fetal MRI data were exported from the hospital PACS (Dedalus HealthCare, Bonn, Germany). Manual segmentation masks of the whole lung (3D ROI) were obtained for initial and repeat T2-weighted axial fetal MRI acquisitions by one radiologist with five years of experience in fetal MRI (F.P.) using ITK-Snap [17]. For each 3D ROI, the slice index at the level of the carina was recorded. MRI images and lung segmentation masks were saved as nifti-files. In addition, image and lung segmentation mask slices at the level of the carina were converted to 2D nifti-files using the python package nibabel (MIT). Radiomics features were extracted from 2 and 3D ROIs using Pyradiomics [18], under Python 3.7.1 with the following settings: normalise parameter 'true', normalise Scale parameter 100, voxelArrayShift 300, (3 SDs × 100) ensuring that only outlier values > 3 SDs below the mean remain negative, binWidth 5, 'sitkBSpline' as interpolator, and resampledPixelSpacing '[2, 2, 2]' for 3D or ‘[2, 2]’ for 2D image data. Radiomics features encompassed the following classes: First Order (n = 18), 3D or 2D Shape (n = 9), Grey Level Co-occurrence Matrix (GLCM, n = 22), Grey Level Size Zone Matrix (GLSZM, n = 16), Grey Level Run Length Matrix (GLRLM, n = 16), and Grey Level Dependence Matrix (GLDM, n = 14). A list of all radiomics features is provided in the Additional file 1. See Fig. 1 for an illustration of the study design.
Study design. Repeated acquisitions of T2-weighted axial MRI images of the lung in a fetus at gestational week 32. Three-dimensional (3D) regions of interest (ROIs) encompassing the whole lung were manually segmented in initial (green) and repeat (blue) acquisitions of 30 fetuses. Two-dimensional (2D) ROIs were defined as lung segmentations at the level of the carina in initial (red) and repeat (orange) acquisitions, respectively. Radiomics feature reproducibility was assessed between features extracted from 2 and 3D ROIs in the initial acquisition (red vs. green), 2D ROIs in repeated acquisitions (red vs. orange), and 3D ROIs in repeated acquisitions (green vs. blue)
Statistical evaluation
R version 4.0.5 (R Core Team, Vienna, Austria) was used for statistical analysis. Intra-class correlation coefficients (ICC) were calculated to assess radiomics feature reproducibility within the initial MRI acquisition between 2 and 3D ROIs, and between initial and repeat MRI acquisition for 2D vs 2D ROIs, and 3D versus 3D ROIs, respectively. The psych R package (version 2.1.9) was used to calculate ICCs, applying two-way mixed effects models (ICC3) and single rater unit. ICCs > 0.9 were considered excellent, > 0.75 to 0.9 good, 0.5 to 0.75 moderate, and < 0.5 poor according to Koo et al. [19]. The paired Wilcoxon Rank Sum test was used to compare ICCs of radiomics feature extracted from 2D ROIs in initial and repeat MRI acquisitions, and of radiomics features extracted from 3D ROIs in initial and repeat MRI acquisitions. Differences in radiomics features between fetuses with pathological or healthy lung development were assessed using ANCOVA followed by estimated marginal mean (EMM, emmeans R package) comparisons, whereby gestational age was included as a covariate and binned into 20 day intervals (< 160, 160–180, 180–200, 200–220 and > 220 days) to control for potential non-linear growth effects. EMM comparison p values were corrected for multiple testing using the false discovery rate (FDR). A two-sided p value of < 0.05 or a false discovery rate of < 0.1 combined with a two-sided p value of < 0.05, as applicable, were considered statistically significant.
Results
Key characteristics of the study cohort are given in Table 1. The median time between initial and repeat axial T2-weighted MRI acquisitions was 3.7 min (IQR 0.9–5.9 min, range 0.7–24.2 min). Radiomics features extracted from 2 and 3D ROIs in the initial MRI acquisition were reproducible to a variable degree (Fig. 2A, B): The median ICC across all features was 0.733 (IQR 0.313–0.814, range 0.018–0.970). Five of 95 (5.3%) features showed excellent, 36 (37.9%) good, 22 (23.2%) moderate, and 32 (33.7%) poor reproducibility. All five features with excellent reproducibility belonged to the class ‘First Order’ (Fig. 2B).
2D versus 3D fetal MRI lung radiomics features. a Intra-class correlation coefficients (ICCs) and 95% confidence intervals (CI) between 2 and 3D ROIs in a single MRI acquisition for each of the 95 investigated radiomics features. Lines indicate ICCs, and ribbons 95% CI. Features are grouped according to class. b Barplots depicting the proportions of radiomics features with poor (ICC < 0.5), moderate (0.5–0.75), good (0.75–0.9) and excellent (> 0.9) reproducibility grouped according to class; CI confidence interval GLCM: grey level co-occurrence matrix, GLDM: grey level dependence matrix, GLRLM: grey level run length matrix, GLSZM: grey level size zone matrix, ICC intra-class correlation coefficient
Radiomics features extracted from 2D ROIs in initial and repeat MRI acquisitions exhibited a median ICC of 0.771 (IQR 0.699–0.835, range 0.048–0.965; see Fig. 3A). Twelve of 95 (12.6%) features showed excellent, 44 (46.3%) good, 26 (27.4%) moderate, 13 (13.7%) poor reproducibility (see Fig. 3B). Radiomics features extracted from 3D ROIs in initial and repeat MRI acquisitions exhibited a median ICC of 0.908 (IQR 0.824–0.929, range 0.335–0.996). Forty-nine of 95 (51.6%) showed excellent, 32 (33.7%) good, 13 (13.7%) moderate, and 1 (1.1%) poor reproducibility (see Fig. 3B). Table 2 shows radiomics feature reproducibility according to feature classes.
Reproducibility of 2D and 3D fetal MRI lung radiomics features. a Intra-class correlation coefficients (ICCs) and 95% confidence intervals (CI) between 2 and 3D regions of interest (ROIs) in repeat examinations for each of the 95 investigated features. Lines (ICCs) and ribbons (95% CI) are coloured according to the comparisons depicted (3D blue, 2D red). Features are grouped according to class. b Barplots depicting the proportions of radiomics features with poor (ICC < 0.5), moderate (0.5–0.75), good (0.75–0.9) and excellent (> 0.9) reproducibility according to feature class and 2D or 3D ROIs in repeat examinations. c Radiomics feature ICCs were significantly higher between initial and repeat MRI acquisitions if 3D ROIs were used for radiomics feature extraction compared to 2D ROIs; CI confidence interval GLCM: grey level co-occurrence matrix, GLDM: grey level dependence matrix, GLRLM: grey level run length matrix, GLSZM: grey level size zone matrix, ICC intra-class correlation coefficient
ICCs were significantly higher between initial and repeat MRI acquisitions if 3D ROIs were used for radiomics feature extraction compared to 2D ROIs (p < 0.001) (see Fig. 3C). A complete list of radiomics feature ICCs between 2 and 3D ROIs in initial MRI acquisition, and between initial and repeat MRI acquisitions using 2D ROIs, and 3D ROIs is provided in the Additional file 1: Table S1.
Means and standard deviations of radiomics features for all fetuses, and for fetuses with normal and pathological lung development, as well as raw and FDR-corrected p values are shown in Additional file 1: Tables S2 (for 3D ROIs) and S3 (for 2D ROIs). Eleven of 100 features (11%) were found to be significantly different between fetuses with normal and pathological lung development when using 3D ROIs (see Additional file 1: Fig. S1). Meanwhile, none of the 100 features (0%) passed the nominal threshold for statistical significance when using 2D ROIs.
Discussion
Radiomics have the potential to enhance the assessment of fetal MRI data by extracting quantitative image features that may not be visually perceivable [20]. However, reliable radiomics-assisted fetal-MRI-based tissue characterisation requires excellent feature reproducibility [11]. The presented findings show that the use of 2D versus 3D lung ROIs for radiomics feature extraction from fetal MRI data severely impacts feature values. In addition, radiomics features extracted from 3D ROIs encompassing the whole fetal lung outperformed features extracted from 2D lung ROIs with regard to reproducibility in repeated image acquisitions. Therefore, in the future, highly-reproducible fetal MRI radiomics features extracted from whole lung segmentation masks may improve non-invasive quantitative assessment of lung development.
Non-invasive in-vivo MRI assessment of the fetal lungs is safe and feasible during the second and third trimesters, which corresponds to the canalicular and saccular phases of lung development [1]. During this time, besides volume growth, lung organogenesis is characterised by microstructural changes including the formation of pulmonary acini, differentiation of type I and II pneumocytes, and increasing production of lung fluid and surfactant. This can be observed in fetal MRI as an increase in lung volume along with an increase in signal intensity [21]. However, visual assessment of fetal lung signal intensity in MRI is subjective. Integration of quantitative measures of tissue characteristics in the form of lung-to-liver, lung-to-muscle, or lung-to spinal fluid signal intensity ratios into fetal MRI lung assessment have produced mixed results [4, 22,23,24], so far prohibiting their translation into clinical routine. Therefore, current image-based assessment of fetal lung development focuses primarily on tissue quantity in the form of lung volume rather than tissue quality. Unfortunately, volume alone is an imperfect descriptor of lung developmental status as gestational age-adjusted growth curves show wide normal ranges [2]. Recently, the use of novel fetal MRI techniques including diffusion-weighted imaging [25], intra-voxel incoherent motion analysis [26] and T2* mapping [27] for the microstructural characterisation of fetal lung tissue has been advocated but their benefit remains largely unclear.
In order to facilitate non-invasive, image-based and timely detection of abnormal fetal lung development, reliable quantitative lung tissue features beyond volume are needed. Radiomics allows the extraction of a multitude of features reflecting various aspects of shape and texture from 2D or 3D image ROIs [28]. Fetal MRI is ideally suited for the extraction of quantitative lung radiomics features since image acquisition follows a standardised protocol. Most fetal imaging centres use a single MRI scanner, which has been shown to be essential for radiomics feature reproducibility. Moreover, dedicated fetal MRI lung assessments already requires manual whole lung segmentation to obtain lung volume [29]. These lung segmentations could be integrated into a post-processing pipeline for radiomics feature extraction that has potential for automation due to the high level of standardisation recommended by the Image Biomarker Standardisation Initiative [30]. Therefore, fetal MRI radiomics analysis of lung development could be implemented without the need for additional costly resources.
Fetal lung texture analysis has thus far only been explored using ultrasound images: Previous studies assessed lung maturity [31], identified cases at risk for pulmonary hypoplasia [32], and predicted neonatal respiratory insufficiency [33] with promising results. However, ultrasound studies used 2D fetal lung ROIs to extract texture features, e.g. from lung tissue at the level of the four chamber view. In addition, physicians placed ultrasound ROIs in ‘representative lung areas’ while avoiding artifacts. This approach raises questions with regard to the reliability of radiomics-based fetal lung assessment: Cardiac position (and the level of the four chamber view) may vary in pathologies where lung development is of particular interest, such as in fetuses with congenital diaphragmatic hernia, or congenital pulmonary airway malformation. Furthermore, it is unclear whether a two-dimensional ROI sufficiently represents tissue characteristics of a much larger three-dimensional structure, particularly if subjective criteria are used to determine the ROI’s location. Lastly, there is a lack of evidence regarding the reproducibility of radiomics features extracted from 2D ROIs in repeated image data acquisitions.
The presented results suggest that radiomics features extracted from 2D fetal MRI lung ROIs do not adequately represent whole lung tissue characteristics. Few features (5 of 95 [5.3%]; 10Percentile, Mean, RootMeanSquared, Median, 90Percentile) showed excellent reproducibility between the use of 2D and 3D ROIs in fetal MRI data from a single acquisition. Moreover, these features all reflected basic first order statistics but none shape or higher-order texture, effectively excluding potentially crucial information contained within the lung’s microstructure from further analysis. Another essential finding concerns the limited radiomics feature reproducibility for the use of 2D lung ROIs in repeated fetal MRI acquisitions. Here, excellent reproducibility was found in only twelve of 95 (12.6%) features. Notably, this was achieved using standardised 2D ROIs that covered the entire lung tissue except hilar structures on an axial fetal MRI slice at the level of the carina. Subjective segmentation of representative-appearing lung areas may have further increased radiomics feature variability. In contrast, excellent radiomics feature reproducibility was found in a majority of features (49 of 95, 51.6%), including a variety of shape and higher-order texture features, if 3D lung ROIs were used for both fetal MRI acquisitions.
The complex and dynamically changing microstructure of the fetal lung are unlikely to be adequately reflected by one quantitative parameter, which may explain the limited utility of previously explored lung signal intensity ratios for outcome prediction [23, 24]. Rather, a combination of images markers, i.e. ‘a radiomics signature’ focusing on different fetal lung characteristics, such as texture and shape, may be more suitable as a quantitative descriptor of lung development. Undoubtedly, lung volume remains an essential parameter to determine whether lung development is age-appropriate. By adding texture and shape features to a lung radiomics signature, the sensitivity of fetal MRI for the detection of abnormal lung development may be increased, e.g. in cases where lung volume is within normal ranges but lung microstructure is altered. As demonstrated, whole lung fetal MRI radiomics enables the extraction of a large number of highly-reproducible lung shape and texture features that may be used to develop radiomics signatures of normal and pathologic lung development in the future.
As a proof of concept, we tested whether 3D and 2D ROIs were sufficient to detect significant differences between radiomics features extracted from fetal lungs with normal or pathological development in our limited sample size. We found 11% of radiomics features were significantly different between lungs with normal compared to pathological development when using 3D ROIs. Critically, in case of the use of 2D ROIs, no significant difference in radiomics features was observed, indicating that radiomics features extracted from 3D ROIs are more sensitive to subtle, visually not perceivable changes in the fetal lung’s microstructure. Further studies are necessary to confirm these findings and identify robust and predictive 3D fetal lung radiomics features.
This study had several limitations: The number of included cases was relatively small but similar or larger compared to previous test–retest studies on radiomics feature reproducibility [13,14,15]. A single 1.5 T MRI scanner was used. Due to the retrospective study design, routinely performed repeated MRI acquisitions from a single examination were utilised rather than repeated fetal MRI scans. While T2-weighted images are widely used for visual assessment of the fetal lung [34], it is not known which MRI sequence is best suited for radiomics-based analysis. The radiomics feature set included in the current analysis is limited, but is compatible with the Image Biomarker Standardisation Initiative guidelines [30]. Pyradiomics has been widely used in lung imaging and beyond facilitating comparability and generalizability of results [18]. Excellent radiomics feature reproducibility was conservatively defined as ICC > 0.9, but there is a lack of evidence concerning an optimal cut-off value.
Conclusion
In conclusion, this study demonstrates that fetal MRI radiomics features extracted from 2D ROIs do not adequately represent tissue characteristics of 3D ROIs encompassing the whole lung. In addition, they exhibit insufficient reproducibility between repeated standardised fetal MRI acquisitions. In contrast, a majority of fetal MRI radiomics features extracted from 3D whole lung segmentation masks are excellently reproducible. Therefore, highly-reproducible whole lung radiomics features represent potential image biomarkers for improved fetal MRI-based prediction of abnormal lung development and neonatal respiratory function.
Availability of data and materials
All data generated or analysed during this study are included in this published article (and its Additional file 1).
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- GLCM:
-
Grey level co-occurrence matrix
- GLDM:
-
Grey level dependence matrix
- GLRLM:
-
Grey level run length matrix
- GLSZM:
-
Grey level size zone matrix
- GW:
-
Gestational weeks plus days
- ICC:
-
Intra-class correlation coefficient
- IQR:
-
Interquartile range
- MRI:
-
Magnetic resonance imaging
- PACS:
-
Picture archiving and communication system
- ROI:
-
Region of interest
References
Kasprian G, Balassy C, Brugger PC, Prayer D (2006) MRI of normal and pathological fetal lung development. Eur J Radiol 57:261–270
Meyers ML, Garcia JR, Blough KL, Zhang W, Cassady CI, Mehollin-Ray AR (2018) Fetal lung volumes by MRI: normal weekly values from 18 through 38 weeks’ gestation. AJR Am J Roentgenol 211:432–438
Cannie MM, Jani JC, Van Kerkhove F et al (2008) Fetal body volume at MR imaging to quantify total fetal lung volume: normal ranges. Radiology 247:197–203
Mills M, Winter TC, Kennedy AM, Woodward PJ (2014) Determination of fetal lung maturity using magnetic resonance imaging signal intensity measurements. Ultrasound Q 30:61–67
Lefebvre TL, Ueno Y, Dohan A et al (2022) Development and validation of multiparametric MRI-based radiomics models for preoperative risk stratification of endometrial cancer. Radiology. https://doi.org/10.1148/radiol.212873:212873
Shin J, Seo N, Baek SE et al (2022) MRI Radiomics model predicts pathologic complete response of rectal cancer following chemoradiotherapy. Radiology 303:351–358
Perez-Johnston R, Araujo-Filho JA, Connolly JG et al (2022) CT-based radiogenomic analysis of clinical stage i lung adenocarcinoma with histopathologic features and oncologic outcomes. Radiology 303:664–672
Du Y, Fang Z, Jiao J et al (2021) Application of ultrasound-based radiomics technology in fetal-lung-texture analysis in pregnancies complicated by gestational diabetes and/or pre-eclampsia. Ultrasound Obstet Gynecol 57:804–812
Jiao J, Du Y, Li X, Guo Y, Ren Y, Wang Y (2022) Prenatal prediction of neonatal respiratory morbidity: a radiomics method based on imbalanced few-shot fetal lung ultrasound images. BMC Med Imaging 22:2
Palacio M, Bonet-Carne E, Cobo T et al (2017) Prediction of neonatal respiratory morbidity by quantitative ultrasound lung texture analysis: a multicenter study. Am J Obstet Gynecol 217(196):e191-196
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Prayer F, Watzenbock ML, Heidinger BH et al (2023) Fetal MRI radiomics: non-invasive and reproducible quantification of human lung maturity. Eur Radiol. https://doi.org/10.1007/s00330-022-09367-1
Prayer F, Hofmanninger J, Weber M et al (2021) Variability of computed tomography radiomics features of fibrosing interstitial lung disease: a test-retest study. Methods 188:98–104
Mitchell-Hay RN, Ahearn TS, Murray AD, Waiter GD (2022) Investigation of the inter- and intrascanner reproducibility and repeatability of radiomics features in T1-weighted brain MRI. J Magn Reson Imaging. https://doi.org/10.1002/jmri.28191
Carbonell G, Kennedy P, Bane O et al (2022) Precision of MRI radiomics features in the liver and hepatocellular carcinoma. Eur Radiol 32:2030–2040
Prayer D, Malinger G, Brugger PC et al (2017) ISUOG Practice Guidelines: performance of fetal magnetic resonance imaging. Ultrasound Obstet Gynecol 49:671–680
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Koo TK, Li MY (2016) A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15:155–163
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Keller TM, Rake A, Michel SC et al (2004) MR assessment of fetal lung development using lung volumes and signal intensities. Eur Radiol 14:984–989
Balassy C, Kasprian G, Brugger PC et al (2010) Assessment of lung development in isolated congenital diaphragmatic hernia using signal intensity ratios on fetal MR imaging. Eur Radiol 20:829–837
Moradi B, Ghorbani Z, Shirazi M et al (2021) Comparison of fetal lung maturation in fetuses with intrauterine growth restriction with control group, using lung volume, lung/liver and lung/muscle signal intensity and apparent diffusion coefficient ratios on different magnetic resonance imaging sequences. J Matern Fetal Neonatal Med. https://doi.org/10.1080/14767058.2021.2008349:1-9
Dutemeyer V, Cordier AG, Cannie MM et al (2022) Prenatal prediction of postnatal survival in fetuses with congenital diaphragmatic hernia using MRI: lung volume measurement, signal intensity ratio, and effect of experience. J Matern Fetal Neonatal Med 35:1036–1044
Afacan O, Gholipour A, Mulkern RV et al (2016) Fetal lung apparent diffusion coefficient measurement using diffusion-weighted MRI at 3 Tesla: Correlation with gestational age. J Magn Reson Imaging 44:1650–1655
Ercolani G, Capuani S, Antonelli A et al (2021) IntraVoxel Incoherent Motion (IVIM) MRI of fetal lung and kidney: Can the perfusion fraction be a marker of normal pulmonary and renal maturation? Eur J Radiol 139:109726
Sethi S, Giza SA, Goldberg E et al (2021) Quantification of 1.5 T T1 and T2 (*) relaxation times of fetal tissues in uncomplicated pregnancies. J Magn Reson Imaging 54:113–121
Mayerhoefer ME, Materka A, Langs G et al (2020) Introduction to radiomics. J Nucl Med 61:488–495
Williams G, Coakley FV, Qayyum A, Farmer DL, Joe BN, Filly RA (2004) Fetal relative lung volume: quantification by using prenatal MR imaging lung volumetry. Radiology 233:457–462
Zwanenburg A, Vallieres M, Abdalah MA et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295:328–338
Zamarian ACP, Caetano ACR, Grohmann RM et al (2022) Prediction of lung maturity in fetuses with growth restriction through quantitative ultrasound analysis. Ultrasound Med Biol 48:20–26
Blitz MJ, Ghorayeb SR, Solmonovich R et al (2021) Fetal lung echo texture in pregnancies at risk for pulmonary hypoplasia. J Ultrasound Med 40:805–810
Burgos-Artizzu XP, Perez-Moreno A, Coronado-Gutierrez D, Gratacos E, Palacio M (2019) Evaluation of an improved tool for non-invasive prediction of neonatal respiratory morbidity based on fully automated fetal lung ultrasound analysis. Sci Rep 9:1950
Avena-Zampieri CL, Hutter J, Rutherford M et al (2022) Assessment of the fetal lungs in utero. Am J Obstet Gynecol MFM. https://doi.org/10.1016/j.ajogmf.2022.100693:100693
Funding
This project has received funding from the Seed Grant funding programme of the European Society of Radiology (ESR) in collaboration with the European Institute for Biomedical Imaging Research (EIBIR) kindly supported by an unrestricted, non-exclusive grant from GE Healthcare.
Author information
Authors and Affiliations
Contributions
MW performed image post-processing including radiomics feature extraction, and formal analysis, and was a major contributor to the writing of the manuscript; BH was a major contributor to the conception of the work and formal analysis; JR performed retrospective review of the hospital PACS and was a major contributor to the writing of the manuscript; VS was a major contributor to the interpretation of the results; BU was a major contributor to the writing of the manuscript; ER was a major contributor to the conception of the study; DP was a major contributor to the conception of the study; GK was a major contributor to the conception of the study, interpretation of results and writing of the manuscript; FP performed the conception of the work, manual segmentation of fetal MRI data, generation of figures, and writing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
This retrospective, single-centre study was approved by the Ethics Committee of the Medical University of Vienna (1232/2022) The requirement to obtain informed consent was waived.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Watzenboeck, M.L., Heidinger, B.H., Rainer, J. et al. Reproducibility of 2D versus 3D radiomics for quantitative assessment of fetal lung development: a retrospective fetal MRI study. Insights Imaging 14, 31 (2023). https://doi.org/10.1186/s13244-023-01376-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13244-023-01376-y