Abstract
Background
Gene silencing by aberrant DNA methylation of promoter regions remains the most dominant phenomenon occurring during tumorigenesis. Improving the early diagnosis, prognosis, and recurrence prediction of colorectal cancer using noninvasive aberrant DNA methylation biomarkers has encouraging potential. The aim of this study is to characterize the DNA methylation of the promoter region of TMEM240, as well as gene expression and its effect on cell biological functions and its applications in early detection and outcome prediction.
Results
Highly methylated CpG sites were identified in the TMEM240 gene by Illumina methylation 450K arrays in 26 Taiwanese patient paired samples and 38 paired samples from The Cancer Genome Atlas (TCGA) colorectal cancer dataset. Transient transfection and knockdown of TMEM240 were performed to demonstrate the role of TMEM240 in colorectal cancer cells. The data showed that TMEM240 could lead to G1 cell cycle arrest, repress cancer cell proliferation, and inhibit cancer cell migration. The quantitative methylation-specific real-time polymerase chain reaction (PCR) results revealed that 87.8% (480 of 547) of the colorectal cancer tumors had hypermethylated TMEM240, and this was also found in benign tubular adenomas (55.6%). Circulating cell-free methylated TMEM240 was detected in 13 of 25 (52.0%) Taiwanese colorectal cancer patients but in fewer (28.6%) healthy controls. In 72.0% (85/118) of tissue samples, TMEM240 mRNA expression was lower in Taiwanese CRC tumor tissues than in normal colorectal tissues according to real-time reverse transcription PCR results, and this was also found in benign tubular adenomas (44.4%). The TMEM240 protein was analyzed in South Korean and Chinese CRC patient samples using immunohistochemistry. The results exhibited low protein expression in 91.7% (100/109) of tumors and 75.0% (24/32) of metastatic tumors but exhibited high expression in 75.0% (6/8) of normal colon tissues. Multivariate Cox proportional hazards regression analysis found that mRNA expression of TMEM240 was significantly associated with overall, cancer-specific, and recurrence-free survival (p = 0.012, 0.007, and 0.022, respectively).
Conclusions
Alterations in TMEM240 are commonly found in Western and Asian populations and can potentially be used for early prediction and as poor prognosis and early-recurrence biomarkers in colorectal cancer.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the most commonly diagnosed cancer in the USA and Taiwan [1, 2]. In the USA and Taiwan, CRC has been the most common cancer for the past 9 years and is now also the third leading cause of cancer deaths in females and males [1,2,3]. CRC is curable in ~ 90% of cases if it is detected at an early stage. The early detection of CRC through screening programs that detect mucosal changes that are predictive of colorectal tumors reduces the incidence and mortality rates of this disease [4]. CRC is a multifactorial disease that arises from an accumulation of acquired genetic and epigenetic alterations in a number of oncogenes, tumor-suppressor genes (TSGs), mismatch-repair genes, and cell cycle genes during tumorigenesis [5,6,7,8]. Among the epigenetic alterations, gene silencing by aberrant DNA methylation of promoter regions remains the most dominant phenomenon occurring during tumorigenesis [9]. Regardless of the biological consequences of methylation-induced silencing of TSGs, this epigenetic alteration constitutes a molecular signature that can serve as a promising biomarker for early detection [8, 10]. Circulating cell-free DNA (ccfDNA) levels reveal a greater dynamic range and have a better correlation with changes in tumor burden than CA-153 or circulating tumor cells [11]. ccfDNA methylation assessment allows for simultaneous screening of CRC complementing current modalities, increasing compliance, and cost-effectiveness [12]. Septin 9 (SEPT9) hypermethylation was previously reported to be used for the early prediction of CRC [13]. SEPT9 methylation in circulating cell-free (ccfDNA) was approved by the US Food and Drug Administration (FDA) as a biomarker for CRC early diagnostic screening. The sensitivity for stages I~III CRC is 64% (48%~77%) [14]. In a Taiwanese CRC cohort, a quantitative polymerase chain reaction (qPCR) revealed that 86.1% of CRC patients exhibited hypermethylated BEND5 [15], suggesting that more sensitive biomarkers were worth investigating in Asian cohorts. In addition, assays of circulating methylated DNA (cmDNA) could be used as outcome predictors in chemotherapy and multikinase inhibitor-treated metastatic CRC patients [16]. These findings encouraged us to identify additional potential CRC-specific methylation markers, and combining multiple biomarkers in the early stage of CRC screening may provide more sensitive CRC early diagnoses [6, 9]. In our investigation, the human Methylation 450K array was used in 26 paired Taiwanese CRC tissues to identify a new potential CRC-specific hypermethylated transmembrane-encoding gene TMEM240.
The TMEM240 gene encodes a transmembrane domain-containing protein found in the brain and cerebellum. Mutations of TMEM240 were found to cause spinocerebellar ataxia 21 (SCA21) with mental retardation, severe cognitive impairment, and hypokinetic and hyperkinetic movement disorders in patients from France, Germany, Holland, Colombia, Japan, and China [17,18,19,20]. The mechanism of the pathogenesis of SCA21 may be mediated through the induction of early gliosis and lysosomal impairment by mutant TMEM240 [21]. Genome-scale analysis of DNA methylation using Infinium HumanMethylation450 BeadChips found hypermethylation of TMEM240 in 22 CRC patients in Russia [22]. However, the role of TMEM240 in tumorigenesis is unclear. Few studies have analyzed the overall methylation status and expression of TMEM240 or its biological functions, clinical significance, or application. Therefore, this study investigated the methylation, expression level, biological functions, clinical significance, and clinical applications of TMEM240 in CRC.
Results
Four potential candidate genes were identified from Taiwanese and Western CRC patients by genome-wide methylation analysis
To identify novel potential biomarkers in CRC patients, we used four criteria to screen potential targets: (1) hypermethylation in Taiwanese CRC patients, (2) a methylation level in normal colorectal tissues of close to 0, (3) hypermethylation in Western CRC patients, and (4) low expression in CRC (Fig. 1a). First, the Illumina Infinium HumanMethylation450 BeadChip array was applied to identify critical TSGs from 26 CRC tissues and paired noncancerous colorectal tissues. In total, 353 genes were found to be hypermethylated when the ΔAvg _β (βTumor–βNormal) was > 0.4. Second, among the 353 genes, 150 genes were selected when the Avg _β value was very low in noncancerous colorectal tissues (Avg _β (Normal) was < 0.1). Next, we analyzed the TCGA Illumina Infinium HumanMethylation450 BeadChip array data of 38 paired Western CRC patients. In total, 1789 genes were found to be hypermethylated when the ΔAvg _β (βTumor–βNormal) was > 0.4. Fourth, the TCGA RNA sequencing data of 41 paired Western CRC patients revealed that the expression levels of 1337 genes were decreased by 80% in CRC. Four candidate genes, ZNF625, TMEM240, SLC6A15, and MPPED2, were identified using InteractiVenn (Fig. 1b). ZNF625, SLC6A15, and MPPED2 have also been found to be hypermethylated in the tumors of CRC patients in previous studies [23,24,25]. MPPED2 was reported to work as a tumor suppressor gene [24, 26, 27]. Few reports about TMEM240 in cancer were found. Analysis of the methylation status in most human digestive systems showed that TMEM240 was frequently hypermethylation in esophageal cancer (12/15, 80.0%), gastric cancer (2/2, 100%), colon cancer (38/38, 100%), rectal cancer (5/5, 100%), liver cancer (37/46, 80.4%), and pancreatic cancer (4/9, 44.4%), compared to paired adjacent normal tissues (Figure S1). Thus, the role of TMEM240 in cancer was unclear. Therefore, TMEM240 in CRC was selected for further analysis. Comprehensive analyses of epigenetic alterations, mRNA and protein expression, and the biological role of TMEM240 were further performed.
TMEM240 protein represses colon cancer cell proliferation and migration
Alterations in TMEM240 and its functional roles during tumorigenesis had not been previously studied. To study the biological roles of the TMEM240 protein in CRC cells, a TMEM240 plasmid or si-TMEM240 was transfected by the electroporation systems into DLD-1 cells which exhibited low level of TMEM240 expression (Figure S2). The gene manipulation efficiency was determined through real-time RT-PCR and Western blotting. Transfection of the TMEM240 plasmid into the DLD-1 cell line resulted in an abundant display of TMEM240 mRNA and protein by DLD-1 cells. According to the cell viability sulforhodamine B (SRB) assay, the growth of DLD-1 cancer cells with TMEM240 was slow down by 66.6% compared to that of the vector control group (Fig. 2a). Microscopic observations revealed that TMEM240 overexpression repressed the growth of DLD-1 cells. To verify whether the decrease in TMEM240 expression induced cell growth, knockdown of TMEM240 gene expression was achieved in the DLD-1 cell line with si-TMEM240. TMEM240 knockdown increased DLD-1 cell growth by 2.0-fold (Fig. 2b). We further transfected with two si-TMEM240 (s50536) and si-TMEM240 (s50534) into the colon cancer cell line HCT116, respectively. TMEM240 mRNA expression was dramatically decreased by two si-TMEM240, s50534 and s50536. The microscopy and SRB assays showed that both si-TMEM240 s50534 and s50536 could significantly induce colon cancer growth by 15.9- and 14.9-fold, respectively (Fig. 2c).
TMEM240 may repress cell growth, migration, and cell cycle arrest in colon cancer cells. a A recombinant pMyc-DDK-hTMEM240 plasmid was transfected into DLD-1 cells for 24 h, and then the cells were analyzed via real-time RT-PCR for mRNA and Western blotting for protein (left). The cell proliferation rate was analyzed by sulforhodamine B (SRB) assay (middle). A bright view was taken to illustrate the cell morphology (right) (original magnification, × 100). b si-TMEM240 was transfected into DLD-1 cells. The mRNA expression (up), cell proliferation rate (down), and cell morphology (right) of DLD-1 cells were analyzed (original magnification, × 100). c The mRNA expression (up), cell morphology (right), and cell proliferation rate (down) in HCT116 colon cancer cells were analyzed (original magnification, × 40). d The migratory ability was measured by a transwell assay after TMEM240 overexpression in DLD-1 cells. e A TMEM240 and/or si-TMEM240 plasmid was transfected into DLD-1 cells for 24 h, and then the cell cycle distribution was analyzed by flow cytometry. Data are presented as the mean ± SD, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. A t test was used to calculate group differences in all experiments. Experiments were performed with at least two biological duplicates and three technical replicates
To investigate whether TMEM240 was associated with colon cancer cell migration, TMEM240 was overexpressed in DLD-1 cells for 24 h. Next, cell motility was analyzed using transwell assays. The data revealed that the increase in TMEM240 suppressed the migratory ability of DLD-1 cells by 39.7% (Fig. 2d).
TMEM240 expression induced cancer cell arrest in the G1 phase
The data of Fig. 2 have indicated that the TMEM240 gene inhibits the growth of cancer cells and that the growth of cells requires regulation of the cell cycle. Therefore, cell cycle regulation by TMEM240 was studied by flow cytometry. The percentage of DLD-1 cells in the G1 phase increased by 4.28% in TMEM240-knockdown cells compared to the vector control. TMEM240 knockdown induced a decrease in the proportion of cells in the G1 phase by 6.33% compared to the siRNA control. An opposite trend was observed for the G2M phase, i.e., the percentage of DLD-1 cells in the G2M phase decreased by 4.53%, and TMEM240-knockdown induced a 2.05% increase in the proportion of cells in the G2M phase compared to the control (Fig. 2e).
Hypermethylation of TMEM240 in CRC tumors and plasma from Taiwanese CRC patients
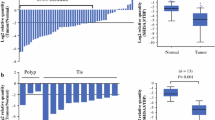
TMEM240 was found to have tumor suppressor potential via affecting CRC cell growth and migration (Fig. 2) and was found to be hypermethylated in both Taiwanese and Western CRC patients (Fig. 1). Therefore, we further comprehensively analyzed DNA methylation alterations and mRNA and protein expression in Asian and Western CRC patients. In 26 paired Taiwanese CRC patients, the Illumina Infinium HumanMethylation450 BeadChip array found three methylation differences in the ΔAvg _β (βTumor–βNormal) between paired CRC tumor and adjacent normal tissues at of the cg16601494, cg15487867, and cg16306898 sites, and the ΔAvg _β values at these sites were 0.47, 0.45, and 0.41, respectively, in the promoter and exon regions of the TMEM240 gene. In the TMEM240 gene, 30 CpG sites were identified, and an increase in the methylation level was found mainly in the promoter and exon 1 regions, most significantly in CpG sites, in CRC tumors compared to adjacent normal tissues, as shown in a heatmap in Fig. 3a.
Methylation levels in Taiwanese colorectal carcinoma (CRC) patients. a Differentially methylated CpG heatmap of TMEM240 in 26 paired CRC patients. Methylation levels (average β values) at differentially methylated loci were identified using an Illumina Methylation 450K array-based assay. The highly differentially methylated sites cg15487867, cg16601494, cg16306898, and cg22497741 are array probes 6, 7, 8, and 9, respectively. The genomic positions of the QMSP amplicon are located in exon 1 (from + 171 to + 347) of the TMEM240 gene (array probes 7 and 8). b The box plot of TMEM240 methylation levels in tissues. c Representative figures of TMEM240 methylation levels determined by QMSP in 10 adjacent normal colon tissues, nine polyps of tubular adenoma, and 10 CRC tumors. Experiments were performed with three technical replicates. d The box plot of TMEM240 methylation levels in plasma. e Representative figures of the circulating methylated TMEM240 levels determined by QMSP in 10 healthy subjects and 10 CRC patients. *p ≤ 0.05, ***p ≤ 0.001. A t test was used to calculate group differences
Methylation patterns of TMEM240 were verified by performing QMSP assays in 547 CRC patients and nine patients with benign tubular adenoma. The median DNA methylation levels that were normalized by ACTB were 0.0098 in CRC tumors, 0.0015 in benign tumors, and 0.0001 in adjacent normal tissues (Fig. 3b, Figure S3, Table 1). The data indicated that in 87.8% (480/547) of Taiwanese CRC patients, the methylation level of TMEM240 was at least 5-fold higher in tumor tissues than in the matched normal colorectal tissues (Fig. 3 b and c, Table 1). Hypermethylation of the TMEM240 promoter was detected in five of nine (55.6%) patients with a benign tubular adenoma (Table 1). DNA hypermethylation of TMEM240 is especially higher in the cecum or appendix and transverse colon than in other locations (Table 1). In addition, in plasma, the median DNA methylation levels were 0.0021 in CRC patients and 0.0000 in healthy controls. The hypermethylation of the TMEM240 promoter was detected in plasma circulating cell-free DNA from 13 of 25 (52.0%) CRC patients, but less TMEM240 promoter hypermethylation was detected in healthy subjects (12 of 42, 28.6%, Fig. 3 d and f).
Low TMEM240 mRNA expression in Taiwanese CRC patients and their associated poor prognoses
We further analyzed TMEM240 mRNA expression in 118 paired CRC tissues. The median TMEM240 mRNA expression levels normalized by GAPDH were 0.0114 in CRC tumors and 0.0485 in adjacent normal tissues. (Fig. 4a, Figure S4, Table 1). In 72.0% (85/118) of tissue samples, TMEM240 mRNA expression was lower in Taiwanese CRC tumor tissues than in normal colorectal tissues (Fig. 4a, Table 1). Loss of mRNA expression is especially higher in the cecum or appendix and ascending colon than in other locations (Table 1). Low TMEM240 mRNA expression was found in four of the nine samples (44.4%) from patients with benign tubular adenoma (Fig. 4b, Table 1g). CRC patients with low mRNA expression of the TMEM240 gene had relatively lower overall survival (OS) and cancer-specific survival (CSS) rates than CRC patients with high mRNA expression of TMEM240 (p = 0.048 and 0.026, respectively) according to Kaplan-Meier survival curves and a log-rank test of 78 Taiwanese CRC patients, and this was especially true in female, stage IV and younger CRC patients (Fig. 4c, Figure S5, p = 0.021, 0.008, and 0.009, respectively). A Cox proportional hazards survival analysis further adjusted for sex, age, tumor stage, tumor location, and differentiation showed that TMEM240 promoter hypermethylation and low mRNA expression were significantly and independently associated with OS, CSS, and recurrence-free survival (RFS) (Table 2, p = 0.028, 0.025, and 0.045, respectively).
TMEM240 mRNA expression levels are affected by DNA methylation and are associated with cancer-specific survival. a The box plot of TMEM240 mRNA expression levels in tissues. b Representative figure of the TMEM240 mRNA expression level determined by RT-qPCR in 10 adjacent normal colon tissues, nine polyps of tubular adenomas, and 10 colorectal carcinoma (CRC) tumors. c Kaplan-Meier survival curves were used to compare cancer-specific survival between CRC patients with low and those with high TMEM240 mRNA expression. TMEM240 was considered to have low expression when the expression level in CRC tumors was 5-fold lower than that in normal tissues. d Scatterplot of the fold change of TMEM240 DNA methylation and mRNA expression levels in CRC tumors normalized to the adjacent normal tissues. e DNA methylation and mRNA expression were measured after treatment with decitabine (DAC). Fold changes in DNA methylation after treatment with DAC (left panel). Fold changes in TMEM240 mRNA expression after treatment with DAC (right panel). Data are presented as the mean ± SD, **p ≤ 0.01, ***p ≤ 0.001. A t test was used to calculate group differences in all experiments. Experiments were performed with at least two biological duplicates and three technical replicates
The DNA hypermethylation levels and mRNA expression levels of TMEM240 were significantly negatively correlated according to a Spearman rank correlation analysis (p = 0.045, Fig. 4d). To determine whether hypermethylation of TMEM240 was associated with mRNA expression, the suppression of TMEM240 mRNA expression by hypermethylation was investigated via administration of the DNA demethylating drug decitabine (DAC) in the DLD-1 colon cancer cell line. Cells were treated with DMSO and DAC for 48 h. In the DAC group, the degree of methylation of TMEM240 decreased to 60.57% of that in the DMSO group (p = 0.001, Fig. 4e), and TMEM240 mRNA expression increased by 2.02-fold (p < 0.001, Fig. 4e), suggesting that hypermethylation of the TMEM240 promoter is the main mechanism of TMEM240 silencing.
Low TMEM240 protein expression in south Korean and Chinese CRC patients
TMEM240 protein expression was analyzed in 109 tumors from CRC patients, 32 CRC metastatic tumors from patients, 10 benign tumors, eight hyperplastic tumors, 10 colon tissues with inflammation, 11 adjacent normal tissues, and eight normal colon tissues by IHC. The small, round, positive cells are lymphocytes, which serve as internal control cells (Fig. 5a). The TMEM240 protein was mainly expressed in the plasma membranes of the cells of the normal colonic mucosa (Fig. 5a). Note that positive TMEM240 protein staining was detected in 87.5% (7/8) and 70% (7/10) of the hyperplastic colon tumors and benign tumors, respectively (Fig. 5 b and c, Table 1). But it was deficiently or weakly expressed in the cytoplasm and nuclei of malignant CRC tumor cells (Fig. 5d–f). The TMEM240 protein level decreased in 91.7% (100/109) of tumors from CRC patients and in 75.0% (24/32) of the metastatic tumors from CRC patients but exhibited higher expression in 75.0% (6/8) of the normal colon tissues (Fig. 5, Table 1). The protein expression of TMEM240 was deficiently especially in the cecum or appendix, ascending, sigmoid colon, and rectum than in other locations (Table 1).
Representative figures for TMEM240 protein expression as analyzed by IHC. a Normal colon mucosa. b Hyperplastic polyp. c Benign colon tumor. d Colon cancer tissue with negative expression. e Colon cancer tissue with low expression. f Metastatic colon cancer tissue (original magnification, × 200). Scale bars indicate 200 μm
TMEM240 promoter hypermethylation and low mRNA expression in the CRC tissues from the TCGA dataset
To further determine alterations in TMEM240 hypermethylation and mRNA expression in Western CRC patients, we first analyzed the TCGA data of 38 CRC tumors, 38 matched normal tissues, and 314 CRC tumor tissues that had been analyzed with an Illumina Infinium HumanMethylation450 BeadChip array and displayed the methylation levels with a heatmap. Again, the exon 1 region of TMEM240 was hypermethylated in 314 colorectal tumor tissues (Fig. 6a, b). Analysis of RNA sequencing data from TCGA showed that TMEM240 mRNA expression was markedly significantly reduced in 41 paired CRC tumor tissues compared to the matched normal colorectal tissues (p = 0.019, Fig. 6c). TMEM240 promoter hypermethylation and low mRNA expression were found in 83.6% (245/293) and 85.3% (389/456) of CRC tumors, respectively (Table S2). The DNA hypermethylation levels and mRNA expression levels of TMEM240 showed a significant negative correlation by Spearman rank correlation analysis (p = 0.045). Hypermethylation of TMEM240 was associated with older CRC patients and patients without the kras mutation (p = 0.008 and 0.038, respectively, Table S2). Low expression of TMEM240 mRNA was associated with CRC patients without regional lymph node metastasis (p = 0.028, Table S2). However, using Kaplan-Meier survival curves and a log-rank test to analyze whether the low expression of TMEM240 is associated with poor survival in the TCGA data, the result shows no significant correlation between low expression of TMEM240 and survival (p = 0.163).
TMEM240 DNA methylation and mRNA analysis from the TCGA dataset. Differentially methylated CpG sites in TMEM240 were identified in a 38 adjacent normal colorectal tissues, 38 matched colorectal carcinoma (CRC) tumors, and b 314 CRC tumors by an Illumina Methylation 450K array-based assay. c RNA sequencing data for TMEM240 in 41 adjacent normal colorectal tissues and 41 matched CRC tumors
Discussion
Aberrant promoter hypermethylation of CpG islands associated with TSGs can cause transcriptional silencing and contribute to tumorigenesis. Advancements in detection technology have reduced CRC death rates in several Western countries [28]. Therefore, developing biomarkers for early detection and intervention can improve patient outcomes. Recent studies reported that several TSGs are often methylated in the tumorigenesis that involves the transformation of normal colonic epithelium to adenocarcinoma [29]. In the present investigation, highly methylated CpG sites in the promoter and exon 1 regions of TMEM240 were identified using genome-wide methylation arrays. A QMSP confirmed TMEM240 hypermethylation in CRC tumor tissues compared to normal tissues. In the Asian and TCGA cohorts, hypermethylation of the promoter region of TMEM240 in more than 80% of tumors was consistently found regardless of age, sex, tumor type, stage, tumor size, regional lymph node metastasis, or distant metastasis. Low expression of the TMEM240 protein was found in most Korean and Chinese CRC patients. Moreover, patients with low TMEM240 mRNA expression had poor OS, CSS, and RFS, especially in young and stage IV CRC patients. We speculate that DNA hypermethylation induces lower expression of TMEM240 and then subsequently induces poor survival. However, the TMEM240 expression was not associated with survival in TCGA data set, although TMEM240 DNA hypermethylation and low mRNA expression were commonly observed in TCGA. Whether low expression of TMEM240 was correlated with survival, only in Asian CRC patients need to be further studied in more specimens from Asian and Western countries using the same experimental technique.
Low expression of the TMEM240 protein was also observed in 50.0% of patients with inflammatory bowel diseases (IBDs) (Table 1). Individuals with IBDs have a 3~5-fold increased risk of CRC, as reported in a previous study [24]. Whether TMEM240 also plays an important role in IBDs during CRC tumorigenesis is worthy of further investigation. Although the anti-TMEM240 antibody used on the Human Protein Atlas website (https://www.proteinatlas.org/ENSG00000205090-TMEM240) is the same as the one we used (Sigma-Aldrich, HPA066721), it does not detect TMEM240 in normal colon tissues but does detect it in Purkinje cells in the human cerebellum. We used normal cerebellum Purkinje cells confirm the positive signal. Because TMEM240 mRNA expression was successfully stably detected at a low level in almost all normal colorectal tissues, we tried to find better antibody conditions for protein detection. Therefore, we also tested the antibody at different dilutions. We found that the staining was obvious in the membrane compartment of normal colorectal tissues when the antibody was diluted at a 1:35 ratio but still exhibited no staining in colorectal tumor parts. The immunoexpression in CRC tissues was also consistent with our mRNA expression results. Therefore, we suggested that a lower dilution could be better for detecting lower levels of TMEM240 protein expression.
The TMEM240 gene encodes a transmembrane domain-containing protein found in the brain and cerebellum. The protein structure in UniProtKB/Swiss-Prot showed that the protein has two transmembrane protein regions located at amino acids 5~25 and 90~110. We observed that the TMEM240 protein was mainly expressed in the plasma membranes of the cells of the normal colon mucosa but was deficiently or weakly expressed in the cytoplasm and nuclei of malignant CRC tumor cells (Fig. 5). The data is similar to the result of mRNA expression level. Whether the distribution of TMEM240 protein across cell compartments, ranging from plasma membranes to the cytoplasm and nuclei, and the accompanying biological functional loss are involved in tumorigenesis needs to be further investigated. The precise protein location will be worth of further investigation. The single-cell RNA-sequencing or single-cell proteomic mass spectrometry approaches would be worth to explore specific colon cellular gene and protein expression and localization [30, 31].
The biological role and function of the TMEM240 protein are unclear. In vitro cell model studies have suggested that both the knockdown and overexpression of TMEM240 can repress cell proliferation. Flow cytometry revealed that TMEM240 knockdown induced a decrease in the proportion of cells in the G1 phase and an increase in cells in the G2/M phase. However, an opposite trend was observed when TMEM240 was overexpressed (Fig. 2). These data suggest that TMEM240 reduces 4.28%~6.33% of the cell proportion through cell cycle arrest and probably participates in other mechanisms, such as apoptosis, autophagy, and cellular differentiation. Whether TMEM240 protein is a potential anticancer target is worthy of investigation. DAC treatment increased TMEM240 mRNA expression through TMEM240 promoter demethylation, suggesting that hypermethylation of the TMEM240 promoter is the principle mechanism behind TMEM240 silencing. In addition, DAC sensitizes CRC cells to topoisomerase inhibitors (irinotecan, etoposide, doxorubicin, and mitoxantrone) [32]. Whether DAC, DNA methyltransferase inhibitors, or other DNA demethylating agents exert relevant anticancer effects through inducing TMEM240 expression is worthy of further investigation [33].
TMEM240 DNA methylation and mRNA expression were analyzed in 556 Taiwanese patients. TMEM240 protein expression was analyzed in 108 South Korean patients and 83 Chinese patients. Our study also analyzed the DNA methylation and mRNA expression of TMEM240 in 456 Western TCGA patients. The results support the idea that hypermethylation and decreased expression of TMEM240 are common characteristics in patients from both Asian and Western countries. Note that hypermethylation of TMEM240 was found in 87.2% of Taiwanese CRC tissues, which is much higher than that found for hypermethylation of SEPT9 (60.92%) in Taiwanese CRC patients. The data suggest that hypermethylation of TMEM240 could be a superior early predictive biomarker in Asian CRC patients. In addition, hypermethylation of TMEM240 was found in 55.6% of patients with polyp tubular adenomas, supporting hypermethylation of TMEM240 as an early-onset indicator of CRC. Although the sensitivity of TMEM240 (52.0%) for stages I~III CRC was less than that of SEPT9 (68%, stages I~III), the sensitivity was analyzed with 3.5 mL of plasma. However, our study was limited by the fact that the available volume of plasma for the free methylated TMEM240 analysis was 200 μL, which is much less than the 3.5 mL of plasma used for the analysis of free methylated SEPT9. Approximately 90% of CRC patients exhibit TMEM240 hypermethylation, and the detection of circulating methylated TMEM240 will still improve early diagnosis and provide a noninvasive method to monitor the residual tumor burden after treatment. Therefore, free methylated TMEM240 is a potential biomarker for the early prediction of CRC, encouraging us to further enroll additional patients and perform a study with more plasma samples. However, circulating methylated TMEM240 showed a 28.6% false-positive rate for early prediction. Improving the specificity of circulating cell-free DNA extraction to decrease residual genomic DNA during extraction may decrease the false-positive rate. The addition of other genes with altered methylation levels, such as ZNF625, TMEM240, SLC6A15, and MPPED2. The ZNF625, SLC6A15, and MPPED2 had been also found hypermethylation in tumors of CRC patients in previous studies [23,24,25]. However, whether the hypermethylation of those genes can be detected in ccfDNA is worth further investigation. In addition, the hypermethylation of the ZNF625, TMEM240, SLC6A15, and MPPED2 genes used in the predictive model may largely increase the sensitivity, improve the accuracy, and decrease the false-positive rate.
Interestingly, TMEM240 hypermethylation was also found in other cancers of the digestive system, such as esophageal and liver cancers. The identification of TMEM240 hypermethylation using Infinium HumanMethylation450 BeadChips was also reported in the US hepatocellular carcinoma patients [34]. We also analyzed other cancers, such as lung cancer, and a lower aberrant frequency was found. These data imply that TMEM240 hypermethylation mainly occurs in the gastrointestinal tract, especially in the regions involved in food transport, including the esophagus, stomach, colon, and rectum. Whether aberrant hypermethylation of TMEM240 is associated with eating habits is also worthy of further study. In addition to its use in CRC, TMEM240 hypermethylation may also be an early-onset indicator in gastrointestinal tract cancer.
Conclusions
The current study revealed that TMEM240 hypermethylation was an early-onset CRC indicator. Thus, circulating free methylated TMEM240 is associated with poor prognosis and can be applied as a noninvasive biomarker for early detection and early recurrence prediction.
Methods
Patients and tissue, plasma collection
In total, 547 human CRC tissues, 547 paired adjacent normal colorectal tissues, nine polyp (tubular adenoma) tissues, and 67 plasma samples were obtained from the Taipei Medical University (TMU) Joint Biobank and Taipei Veterans General Hospital Biobank (Fig. 7). Before clinical data and sample collection, written informed consent was obtained from all patients. Patients undergoing preoperative chemoradiotherapy or an emergent operative procedure, those who died within 30 postoperative days, or those with evidence of familial adenomatous polyposis or Lynch syndrome were excluded from this study. Sections of cancerous tissues and corresponding noncancerous tissues were reviewed by a senior gastrointestinal pathologist. The adjacent nontumor part was collected from the colonic mucosa 10 cm proximal from the main tumor. Clinical data on sex, personal and family medical histories, tumor location, TNM tumor stage, tumor differentiation, microsatellite instability (MSI), pathological features, and follow-up conditions, which were prospectively collected, were obtained from the TMU Joint Biobank and Taipei Veterans General Hospital Biobank. Following surgery, patients were monitored every 3 months for the first 2 years and semiannually thereafter.
Genomic DNA, circulating cell-free (cf) DNA, and RNA extraction
Genomic DNA from matched pairs of primary tumor tissues and adjacent colorectal tissues from the same patient was extracted using the QIAamp DNA Mini Kit (Qiagen, Bonn, Germany, cat. no. 51306) according to the manufacturer’s instructions. For RNA extraction, tumor and normal specimens were immediately frozen after surgical resection and stored at − 80 °C. Total mRNA was extracted using the RNeasy Plus Mini Kit (Qiagen, cat. no. 74134) according to the manufacturer’s instructions. ccfDNA was extracted from plasma. The ccfDNA of 26 plasma samples was extracted using a MagMAX Cell-Free DNA Isolation Kit according to the manufacturer’s recommended protocol (Thermo Fisher Scientific, Austin, TX, USA) [35, 36]. The ccfDNA in 200 μL of plasma for each of the 41 patients was extracted using a QIAamp MinElute Virus Spin Kit according to the manufacturer’s recommended protocol [37].
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
To measure TMEM240 mRNA expression, real-time RT-PCR was performed with a LightCycler 480 (Roche Applied Science, Mannheim, Germany). Real-time PCR was performed using the LightCycler 480 Probe Master Kit (Roche Applied Science, Indianapolis, IN, USA, cat. no. 04707494001) with specific primers and a corresponding universal library probe (Roche Applied Science) according to the manufacturer’s instructions. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as a reference gene. PCR conditions were as follows: preincubation, 95 °C for 10 min; and amplification, 95 °C for 10 min and 60 °C for 10 min, for a total of 45 cycles. Normalized gene expression values obtained using the LightCycler Relative Quantification software (vers. 1.5, Roche Applied Science) were then compared to those of the control group. TMEM240 mRNA expression was considered low if the mRNA expression level of TMEM240 relative to GAPDH was 0.5-fold lower in colorectal tumor tissues than in paired normal colorectal tissues. The primers and probes used in the RT-PCR are listed in Supplementary Table S1.
TaqMan quantitative methylation-specific PCR (QMSP)
After bisulfite conversion of genomic DNA using the EpiTect Fast DNA Bisulfite Kit (Qiagen, Bonn, Germany, cat. no. 59826) according to the manufacturer’s recommended protocol, the DNA methylation level of TMEM240 was measured using TaqMan QMSP with a LightCycler 480 (Roche Applied Science). The QMSP was performed with the SensiFAST™ Probe No-ROX Kit (Bioline, London, UK, cat. no. BIO-86020) with specific primers and a methyl-TaqMan probe for TMEM240. Normalized DNA methylation values, which were calibrated to the control group, were obtained using the LightCycler Relative Quantification software (vers. 1.5, Roche Applied Science). The beta-actin (ACTB) gene was used as a reference gene. Primers and probes for TMEM240 methylation detection were designed for the junction between the promoter and exon 1 regions. The relative TMEM240 DNA methylation level was determined with a methylated specific TMEM240 probe and primer (with 7 CpG sites) and normalized to ACTB (without a CpG site). ACTB can work as total genomic DNA content control. Only when all 7 CpGs of TMEM240 genes show methylation at the same time can the PCR be successfully amplified and detected. In 7 total CpG sites, 3 CpGs were detected by forward primer, one CpG was detected by probe, and 3 CpGs were detected by reverse primer. TMEM240 was considered hypermethylated when the methylation level of TMEM240 relative to that of the ACTB gene was at least 2-fold higher in the colorectal tumor compared to the paired normal colorectal tissue sample. The specificity of TMEM240 methylation end products was confirmed by bisulfite sequencing. The primers and probes used in the QMSP are listed in Supplementary Table S1.
Genome-wide methylation analysis
The genome-wide methylation analysis of 26 paired CRC tissues and corresponding noncancerous colon tissues was performed using the Illumina Infinium HumanMethylation450 BeadChip array (Illumina, San Diego, CA, USA), as previously described [15, 17]. The array contains more than 450,000 methylation sites and provides genome-wide coverage of gene regions and CpG island coverage, including 99% of RefSeq genes. Bisulfite conversion was performed for 500 ng of genomic DNA using the EpiTect Fast DNA Bisulfite Kit (Qiagen, cat. no. 59826), according to the manufacturer’s instructions. Methylation scores for each CpG site are represented as “beta” values ranging from 0 (unmethylated) to 1 (fully methylated), which were calculated by determining the ratios of methylated signal intensities to the sums of the methylated and unmethylated signal outputs.
Cell lines, cell culture, and drug treatment
The DLD-1, HCT116, and colo320-DM cell lines used in this study are CRC cells obtained from the Bioresource Collection and Research Center (http://www.bcrc.firdi.org.tw/). The DLD-1 and colo320-DM were cultured in UltraCulture serum-free medium (Lonza, Walkersville, MD, USA, cat. no. 12–725F). HCT116 cells were cultured with 2.5% human platelet lysate (hPL, Compass Biomedical, MA, USA) and 1% penicillin/streptomycin (Thermo Fisher Scientific). To demethylate TMEM240, DLD-1 cells were treated with dimethyl sulfoxide (DMSO) or the demethylation agent decitabine (DAC, Sigma-Aldrich, St. Louis, MO, USA). After treatment, DNA and RNA were extracted, and methylation and expression levels were analyzed. DAC was dissolved in DMSO.
Immunohistochemistry (IHC) assay
Three sets of tissue microarrays of colon tissues and CRC tissues were created to analyze TMEM240 protein expression. Two sets of tissue microarrays of CRC were purchased from SuperBioChips Laboratories (cat. nos. CD4 and CDA3; Seoul, South Korea). A third tissue microarray was purchased from Biomax (cat. no. CO2081a; US Biomax, Rockville, MD, USA). Tissue microarrays were composed of colorectal tumor tissues obtained from 105 South Korean samples of CRC tumors and 104 Chinese samples, including 45 CRC tumors, 32 CRC metastatic tumors, 10 benign tumors, 8 hyperplastic tumors, 10 colon tissues with inflammation, 11 adjacent normal tissues, and 8 normal colon tissues (Fig. 7). The pathologic diagnoses of these cases were microscopically confirmed by two researchers. IHC staining with an anti-TMEM240 antibody (1:35, Sigma-Aldrich, HPA066721) was performed using an iView DAB detection kit (Ventana, Tucson, AR, USA) on a BenchMark XT autostainer. This assay included both positive and negative controls. The researchers who evaluated the IHC staining results were blinded to the clinical follow-up data. The intensity of TMEM240 expression was identified semiquantitatively as no expression, low expression (weaker than or equal to the expression intensity of normal colon epithelium), or high expression (stronger than the expression intensity of normal colon epithelium).
Flowchart of the study design, datasets, and specimens used. For each step, the sample types and number of samples used for the analyses are indicated. CRC, colorectal cancer; AN, adjacent normal; AD, benign adenoma; inflam, inflammation; normal, normal tissues; ccfDNA, circulating cell-free DNA; QMSP, quantitative methylation-specific PCR; qRT-PCR, quantitative reverse-transcription PCR; IHC, immunohistochemistry; methylation450K array, Illumina Infinium HumanMethylation450 BeadChip array; OS, overall survival; CS, cancer-specific survival; RFS, recurrence-free survival; SRB, sulforhodamine B assay; PI, propidium iodide staining
Immunoblotting analyses
For Western blotting assays, the cells were lysed on ice in radioimmunoprecipitation buffer (0.05 M Tris-HCl [pH 7.4], 0.15 M NaCl, 0.25% deoxycholic acid, 1% IGEPAL CA-630, and 1 mM ethylenediaminetetraacetic acid). The lysates were centrifuged at 13,000 rpm at 4 °C for 10 min. The protein extracts were solubilized in sodium dodecyl sulfate (SDS) gel loading buffer (60 mmol/L Tris base, 2% SDS, 10% glycerol, and 5% β-mercaptoethanol). Samples containing equal amounts of protein (40 μg) were separated on an 8% SDS-polyacrylamide gel by using electrophoresis and electroblotted onto Immobilon-P membranes (Millipore, Bedford, MA, USA) in transfer buffer. Immunoblotting was performed using an anti-DDK monoclonal antibody (1:3,000, OriGene, Rockville, MD, US) that was raised against the Myc-DDK-hTMEM240 protein. β-actin (1:5,000, GeneTex, Texas, USA) was used as an internal control.
Plasmid extraction, confirmation, and purification
Plasmid DNA was extracted using a Geneaid™ Midi Plasmid Kit (Geneaid Biotech, cat. no. PI025) according to the manufacturer’s instructions. Plasmid DNA was then subjected to a preliminary length analysis by restriction enzyme digestion and sequenced to confirm accurate production. The plasmid concentration was measured with a NanoDrop 2000C ultra-microwave-length spectrometer (Thermo Fisher Scientific, Wilmington, DE, USA) and stored at − 20 °C until further usage.
cDNA expression construct, RNAi, and transfection
The pMyc-DDK-hTMEM240 and pMyc-DDK vector control were used to transfect DLD-1 cells by the Neon® electrotransfection systems and reagent sets (Thermo Fisher Scientific, Cat. No. MPK1025) according to the manufacturer’s instructions. Briefly, 5.6 × 106 cells were seeded and transfected with 1 μg of plasmid at 1100 V for 20 ms. Immediately, cells were transferred to culture medium and incubated. The two si-TMEM240 RNAi molecules that target exon 4 (s50534) and exon 3 (s50536) were obtained from Thermo Fisher Scientific (Cat. No. 4392420).
Cell cycle distribution assay
The cell cycle distribution was studied by flow cytometry. DLD-1 cells (106) were trypsinized and fixed overnight with 80% ethanol at − 20 °C. Fixed cells were stained with a solution containing 20 μg/mL of propidium iodide, 200 μg/mL of RNase A, and 0.1% Triton X-100 for 30 min in the dark. Cell cycle distributions were studied with a FACSCanto II flow cytometer (BD Biosciences, San Jose, CA, USA), and calculations were performed using the ModFIT LT vers. 2.0 software (Verity Software House, USA).
Transwell assay
A transwell assay was used to study cell migration. The transwell separates the upper and lower wells with a semipermeable membrane (Falcon) with a pore size of 8 μm. Approximately, 2 × 104 treated and untreated DLD-1 cells were seeded into the upper layer; 300 μL of serum-free Dulbecco’s Modified Eagle Medium (DMEM)/F12 was added as culture medium, and 800 μL of the serum-containing culture medium was added as a chemical attractant to the lower layer. After 16 h of incubation, cells that had passed through the membrane were washed twice with phosphate-buffered saline (PBS), fixed, and stained with 1% crystal violet/double-distilled (dd) H2O for 60 min at room temperature. Five random areas were photographed with a microscope (Nikon, TokyoJapan), followed by analysis with the ImageJ software to quantify the number of cells.
Sulforhodamine B (SRB) assay
An SRB assay was used to determine the cell growth rate. DLD-1 cells were seeded in 96-well plates at a density of 8000 cells/well and incubated for 24 h. Cells were fixed with 10% trichloroacetic acid for 10 min. After staining with SRB for 30 min, excess dye was removed by washing the cells five times with 1% acetic acid. Cell growth was assessed using a microplate reader to determine the absorbance of the SRB solution at 515 nm. Growth inhibition rates were calculated as follows:
Cell growth inhibition rate (%) = 100 − [(Ti − Tz)/(C − Tz)] × 100 (Ti ≥ Tz);
where Ti = OD of the inhibitor sample, Tz = OD of basal cells, and C = OD of the control. Images were acquired using an inverted microscope (EVOS, AMG, USA) at the indicated time points. Cells were counted using the ImageJ software.
Statistical analysis
Pearson’s X2 test was used to analyze TMEM240 hypermethylation, mRNA, and protein expression in patients with colorectal cancer, and correlations with various clinical parameters, including age, sex, cancer type, stage, degree of differentiation, and degree of infringement, were assessed. All statistical analyses were performed using the SPSS software (SPSS, Chicago, IL, USA). Overall survival, cancer-specific survival, and recurrence-free survival curves were calculated and analyzed using the Kaplan-Meier method and multivariate Cox proportional hazards regression analysis. Comparisons of the survival curves with a log-rank test that resulted in a p of < 0.05 were considered statistically significant.
Availability of data and materials
The data generated in this study are available from the corresponding author upon reasonable request.
Abbreviations
- ACTB:
-
Beta-actin
- ccfDNA:
-
Circulating cell-free DNA
- cmDNA:
-
Circulating methylated DNA
- CRC:
-
Colorectal cancer
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- OS:
-
Overall survival
- QMSP:
-
Quantitative methylation-specific polymerase chain reaction
- RT-qPCR:
-
Reverse-transcription quantitative polymerase chain reaction
- SCA21:
-
Spinocerebellar ataxia 21
- SEPT9:
-
Septin 9
- SRB:
-
Sulforhodamine B
- TCGA:
-
The Cancer Genome Atlas
- TMEM240:
-
Transmembrane protein 240
- TSG:
-
Tumor suppressor gene
- TMU:
-
Taipei Medical University
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Taiwan Ministry of Health and Welfare. 2018 Taiwan health and welfare report. 2019.
Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin D, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer; 2013. Available from: http://globocan.iarc.fr, accessed on 13/12/2013.
Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–76.
Sameer AS, Nissar S. Epigenetics in diagnosis of colorectal cancer. Molecular biology research communications. 2016;5:49–57.
Verma M, Kumar V. Epigenetic biomarkers in colorectal cancer. Molecular diagnosis & therapy. 2017;21:153–65.
Migheli F, Migliore L. Epigenetics of colorectal cancer. Clin Genet. 2012;81:312–8.
Pfeifer GP. Defining driver DNA methylation changes in human cancer. Int J Mol Sci. 2018;19.
Werner RJ, Kelly AD, Issa JJ. Epigenetics and precision oncology. Cancer J (Sudbury, Mass). 2017;23:262–9.
Uehiro N, Sato F, Pu F, Tanaka S, Kawashima M, Kawaguchi K, et al. Circulating cell-free DNA-based epigenetic assay can detect early breast cancer. Breast Cancer Res. 2016;18:129.
Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–209.
Nunes SP, Moreira-Barbosa C, Salta S, Palma de Sousa S, Pousa I, Oliveira J, Soares M, Rego L, Dias T, Rodrigues J et al. Cell-free DNA methylation of selected genes allows for early detection of the major cancers in women. Cancers. 2018;10.
Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–25.
Potter NT, Hurban P, White MN, Whitlock KD, Lofton-Day CE, Tetzner R, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183–91.
Lin RK, Hung WY, Huang YF, Chang YJ, Lin CH, Chen WY, et al. Hypermethylation of BEND5 contributes to cell proliferation and is a prognostic marker of colorectal cancer. Oncotarget. 2017;8:113431–43.
Amatu A, Schirripa M, Tosi F, Lonardi S, Bencardino K, Bonazzina E, et al. High circulating methylated DNA is a negative predictive and prognostic marker in metastatic colorectal cancer patients treated with regorafenib. Front Oncol. 2019;9:622.
Delplanque J, Devos D, Huin V, Genet A, Sand O, Moreau C, et al. TMEM240 mutations cause spinocerebellar ataxia 21 with mental retardation and severe cognitive impairment. Brain. 2014;137:2657–63.
Traschutz A, van Gaalen J, Oosterloo M, Vreeburg M, Kamsteeg EJ, Deininger N, et al. The movement disorder spectrum of SCA21 (ATX-TMEM240): 3 novel families and systematic review of the literature. Parkinsonism Relat Disord. 2019;62:215–20.
Yahikozawa H, Miyatake S, Sakai T, Uehara T, Yamada M, Hanyu N, et al. A Japanese family of spinocerebellar ataxia type 21: clinical and neuropathological studies. Cerebellum (London, England). 2018;17:525–30.
Zeng S, Zeng J, He M, Zeng X, Zhou Y, Liu Z, et al. Spinocerebellar ataxia type 21 exists in the Chinese Han population. Sci Rep. 2016;6:19897.
Seki T, Sato M, Kibe Y, Ohta T, Oshima M, Konno A, et al. Lysosomal dysfunction and early glial activation are involved in the pathogenesis of spinocerebellar ataxia type 21 caused by mutant transmembrane protein 240. Neurobiol Dis. 2018;120:34–50.
Naumov VA, Generozov EV, Zaharjevskaya NB, Matushkina DS, Larin AK, Chernyshov SV, et al. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics. 2013;8:921–34.
Mitchell SM, Ross JP, Drew HR, Ho T, Brown GS, Saunders NF, et al. A panel of genes methylated with high frequency in colorectal cancer. BMC Cancer. 2014;14:54.
Gu S, Lin S, Ye D, Qian S, Jiang D, Zhang X, et al. Genome-wide methylation profiling identified novel differentially hypermethylated biomarker MPPED2 in colorectal cancer. Clin Epigenetics. 2019;11:41.
Lin PC, Lin JK, Lin CH, Lin HH, Yang SH, Jiang JK, et al. Clinical relevance of plasma DNA methylation in colorectal cancer patients identified by using a genome-wide high-resolution array. Ann Surg Oncol. 2015;22(Suppl 3):S1419–27.
Sepe R, Pellecchia S, Serra P, D'Angelo D, Federico A, Raia M, Cortez Cardoso Penha R, Decaussin-Petrucci M, Del Vecchio L, Fusco A et al. The long non-coding RNA RP5-1024C24.1 and its associated-gene MPPED2 are down-regulated in human thyroid neoplasias and act as tumour suppressors. Cancers. 2018;10.
Liguori L, Andolfo I, de Antonellis P, Aglio V, di Dato V, Marino N, et al. The metallophosphodiesterase Mpped2 impairs tumorigenesis in neuroblastoma. Cell Cycle. 2012;11:569–81.
Edwards BK, Ward E, Kohler BA, Eheman C, Zauber AG, Anderson RN, et al. Annual report to the nation on the status of cancer, 1975-2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116:544–73.
Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014;455:43–57.
Abouleila Y, Onidani K, Ali A, Shoji H, Kawai T, Lim CT, et al. Live single cell mass spectrometry reveals cancer-specific metabolic profiles of circulating tumor cells. Cancer Sci. 2019;110:697–706.
Gao S, Yan L, Wang R, Li J, Yong J, Zhou X, et al. Tracing the temporal-spatial transcriptome landscapes of the human fetal digestive tract using single-cell RNA-sequencing. Nature Cell Biol. 2018;20:721–34.
Pawlak A, Ziolo E, Fiedorowicz A, Fidyt K, Strzadala L, Kalas W. Long-lasting reduction in clonogenic potential of colorectal cancer cells by sequential treatments with 5-azanucleosides and topoisomerase inhibitors. BMC Cancer. 2016;16:893.
Tse JWT, Jenkins LJ, Chionh F, Mariadason JM. Aberrant DNA methylation in colorectal cancer: what should we target? Trends in cancer. 2017;3:698–712.
Shen J, Wang S, Zhang YJ, Wu HC, Kibriya MG, Jasmine F, et al. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics. 2013;8:34–43.
Yan L, Chen Y, Zhou J, Zhao H, Zhang H, Wang G. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis. 2018;67:92–7.
Hung CS, Wang SC, Yen YT, Lee TH, Wen WC, Lin RK. Hypermethylation of CCND2 in lung and breast cancer is a potential biomarker and drug target. Int J Mol Sci. 2018;19.
Yuan H, Zhu ZZ, Lu Y, Liu F, Zhang W, Huang G, et al. A modified extraction method of circulating free DNA for epidermal growth factor receptor mutation analysis. Yonsei Med J. 2012;53:132–7.
Acknowledgements
We thank the Core Facility Center of Taipei Medical University for providing the LightCycler 480 system (Roche Applied Science), FACSCanto II flow cytometer (BD Biosciences), ModFIT LT Version 2.0 software (Verity Software House), and Olympus IX71 Inverted Microscope System.
Funding
This work was supported in part by MOST104-2320-B-038-051-MY3 and MOST108-2320-B-038-020 from the Ministry of Science and Technology (Republic of China) and 108-5804-001-400 from the Ministry of Education (Republic of China).
Author information
Authors and Affiliations
Contributions
SCC and RKL designed the studies. SCC and CSH collected and provided the clinical samples. PLL, MA, SLY, SCW, and JYC performed the experiments. RKL, PLL, MA, SCW, JYC, and CSH analyzed and interpreted the data. RKL and SJ drafted the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Taipei Medical University-Joint Institutional Review Board and the Institutional Review Board, Taipei Veterans General Hospital. Written informed consent was obtained from all patients.
Consent for publication
Not applicable
Competing interests
There are no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
Differentially methylated CpG heatmap of TMEM240 in paired colon cancer, esophageal cancer, gastric cancer, pancreatic cancer, rectal cancer, and liver cancer tissues. Methylation levels (average β values) at differentially methylated loci were identified using an Illumina Methylation 450K array-based assay. Figure S2. TMEM240 mRNA expression was analyzed in two colon cancer cell lines and two human normal colon tissues. Figure S3. Representative figures of TMEM240 methylation levels determined by QMSP in 78 adjacent normal colon tissues and 78 CRC tumors. Experiments were performed with three technical replicates. Figure S4. Representative figures of TMEM240 mRNA expression determined by gene specific probe based real-time RT-PCR in 21 adjacent normal colon tissues and 21 CRC tumors. Experiments were performed with three technical replicates. Data are presented as the mean ± SD, * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001. A t-test was used to calculate group differences in all experiments. Experiments were performed with at least two biological duplicates and three technical replicates. Figure S5. The Kaplan-Meier survival curves for (A) overall survival in Taiwanese colorectal carcinoma (CRC) patients and (B) cancer-specific survival in Taiwanese females are shown. TMEM240 was considered to have low expression when the expression level in CRC tumors was 5-fold lower than that in normal tissues. Table S1. TMEM240 mRNA expression and promoter hypermethylation in relation to the clinical parameters of the TCGA CRC dataset. Table S2: List of primers sequences and their reaction conditions used in the present study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chang, SC., Liew, PL., Ansar, M. et al. Hypermethylation and decreased expression of TMEM240 are potential early-onset biomarkers for colorectal cancer detection, poor prognosis, and early recurrence prediction. Clin Epigenet 12, 67 (2020). https://doi.org/10.1186/s13148-020-00855-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-020-00855-z