Abstract
Introduction
Systemic lupus erythematosus (SLE) is a multisystem autoimmune disease. Currently, numerous genetic loci of SLE have been confirmed. Here we try to further explore additional genes contributing to SLE susceptibility in this study.
Methods
Forty nine single nucleotide polymorphisms (SNPs) with moderate-risk for SLE in previous study were genotyped in a large-scale replication study with a total of 3,522 cases and 8,252 controls using the Sequenom Massarray system. Association analyses were performed using logistic regression with gender or sample cohorts as a covariate through PLINK 1.07 software.
Results
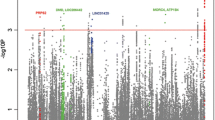
This replication effort confirmed five reported SLE susceptibility loci reaching genome-wide levels of significance (P meta <5.00 × 10−08): TNFSF4 (rs1418190, odds ratio (OR) = 0.81, P meta = 1.08 × 10−08; rs4916219, OR = 0.80, P meta = 7.77 × 10−09), IRF8 (rs2934498, OR = 1.25, P meta = 4.97 × 10−09), miR-146a (rs2431697, OR = 0.69, P meta = 1.15 × 10−22), CD44 (rs2732547, OR = 0.82, P meta = 1.55 × 10−11), and TMEM39A (rs12494314, OR = 0.84, P meta = 1.01 × 10−09). Further logistic regression analysis indicated that the genetic effects within TNFSF4 detected in this study are independent from our previously reported signals.
Conclusions
This study increases the number of established susceptibility loci for SLE in Han Chinese population and highlights the contribution of multiple variants of modest effect. Although further studies will be required to identify the causal alleles within these loci, the findings make a significant step forward in our understanding of the genetic contribution to SLE in Chinese population.
Similar content being viewed by others
Introduction
Systemic lupus erythematosus (SLE) is an important model of autoimmunity characterized by the presence of antibodies to nuclear self-antigens and involvement of multiple organs. The etiopathogenesis of SLE is complex and still largely unknown. Both genetic and environmental factors contribute to disease susceptibility [1]. The prevalence of SLE and its manifestations varies between different ethnic and geographical populations [2], with higher prevalence rates and more severe complications in non-European populations. For example, higher rates of lupus nephritis have been observed in Asians [3,4], Hispanics [5], and African Americans [5,6]. In contrast, higher prevalence rates of photosensitivity have been noted in Europeans [7]. The ethnic and genetic heterogeneity of SLE may contribute to these differences in SLE manifestation rates.
Despite considerable clinical heterogeneity, SLE is one of the most heritable autoimmune diseases, with a sibling risk ratio of around 30 [8]. Increased understanding of the underlying genetic basis for SLE is of key importance in improving the prognosis of patients with SLE. Recent genome-wide association studies (GWAS) and candidate gene studies have confirmed genetic associations of over 40 loci with SLE risk that achieve genome-wide significance (P <5 × 10−8) [9]. Clustering of some genetic associations identified to date appears to fall into at least three major pathways, including type I interferon (IFN) and NF-κB pathways, lymphocyte signaling, and immune complex processing [10]. All of these pathways are of potential importance in the pathogenesis of SLE. However, these genetic risk loci cannot fully explain the genetic susceptibility to SLE and some clinical features, suggesting additional genetic factors yet to be discovered. To detect more additional SLE risk loci, we selected 49 single nucleotide polymorphisms (SNPs) from 40 distinct loci that showed nominal evidence of association to SLE (P <0.01) in our genome-wide dataset [11] and followed up two replications in two large independent sample cohorts of Han Chinese.
Materials and methods
Sample collection
A total of 4,556 SLE patients and 9,451 controls recruited from multiple cooperation hospitals in China were included in this study. The sample information is summarized in Table 1.We included original data from the 1,047 cases and 1,205 controls in the initial stage [11] and the replication samples consisted of two independent cohorts: replication 1 (2,202 SLE cases and 2,208 healthy controls) and replication 2 (1,307 cases and 6,038 healthy controls) from the Han Chinese population. All samples were of self-described Han Chinese descent and cases were confirmed as having SLE by using the revised criteria for the classification of SLE from the American College of Rheumatology (ACR) [12]. Clinical data were collected at each hospital from the affected individuals through a full clinical checkup by at least two physician specialists. Additional demographic information was collected from both cases and controls through a structured questionnaire. The controls’ samples were clinically assessed to be without SLE, other autoimmune disorders, systemic disorders or family history of autoimmune disorders (including first-, second- and third-degree relatives). The Institutional Ethical Committee of The First Affiliated Hospital of Anhui Medical University, according to Declaration of Helsinki principles, approved this study, and all participants provided written informed consent.
EDTA anti-coagulated venous blood samples were collected from all participants in the study. We isolated genomic DNA from peripheral blood lymphocytes by standard procedures using Flexi Gene DNA kits (QIAGEN GmbH, Hilden, Germany). DNA was normalized to working concentrations of 50 ng/μl for genome-wide genotyping and 15 to 20 ng/μl for the validation study.
Genotyping and quality control
Genotyping of the whole genomic was performed using Illumina Human 610-Quad BeadChips (Illumina, Inc., San Diego, CA, USA) in the State Key Lab Incubation of Dermatology, Ministry of Science and Technology, Anhui Medical University (AHMU). We excluded SNPs that had a low call rate (<98%), deviated from Hardy-Weinberg equilibrium (HWE) (P HWE <0.0001 in controls), or the minor allele frequency (MAF) was less than 5% both in cases and controls. The validation study was conducted using the Sequenom Massarray system (Sequenom, Inc., San Diego, CA, USA). Allele detection was performed using MALDI-TOF mass spectroscopy. The mass spectrograms were analyzed by the MassARRAY TYPER software (Sequenom, Inc.). All of these experiments were performed according to the manufacturers’ protocols. Quality control was conducted on each dataset separately using PLINK 1.07 software [13]. According to quality-control measures, we excluded samples with call rates <90% and SNPs with call rates <95%, MAF (<5%) or HWE in controls with P <0.01. In total, 2,202 cases, 2,208 controls and 45 SNPs for replication 1, and 1,307 cases, 6,038 controls and 12 SNPs for replication 2 were available for each replication analysis.
SNPs selection for replication
To search for additional variants associated with SLE, we selected 199 SNPs for further evaluation from our genome-wide dataset after excluding previously reported SLE risk alleles in our studies [11,14,15]. All of these selected SNPs met the following quality criteria: 1) the MAF was higher than 5% both in cases and controls; 2) HWE in controls with P ≥0.01 and HWE in cases with P >0.0001; 3) SNPs with P <0.01 after adjustment by gender; 4) proximity to putative candidate genes (immune-related or involved in immune cell proliferation and differentiation) or known susceptibility loci for autoimmune diseases; and 5) SNPs that appeared to have better evidence of association after further analysis through increasing the sample by another 489 controls. These controls were newly genotyped in a series of GWAS of various diseases in the Han Chinese population, including psoriasis [16], vitiligo [17], leprosy [18], atopic dermatitis [19], esophageal squamous cell carcinoma [20], and so on. However, these additional controls were not well-marched with cases. Therefore, these 489 controls were not included in the association analysis of the validation study or in the final joint association analysis of the combined samples. In each locus, one or two of the most significant SNPs were selected for the validation study. After pruning, 49 SNPs were selected for the replication 1.
Statistical analysis
In the initial stage, we performed single-marker association analyses by logistic regression with gender and age as covariates in the GWAS dataset. Quality control has been described above. For the validation studies, HWE tests in controls were calculated using PLINK 1.07 software. We further excluded SNPs with a call rate of <95% in cases or controls and with HWE in the controls (P <0.001). Additionally, cluster patterns of the genotyping data from the Illumina and Sequenom analyses were checked to confirm their quality. We performed heterogeneity tests (I 2 and P-values of the Q statistics) among these three independent cohorts using the method described previously [21] and the extent of heterogeneity was assessed by using the I 2 index. [22] The fixed effect model (Mantel-Haenszel) was when I 2 was <30% that was considered as no heterogeneity [23]. Otherwise, the random effect model (DerSimonian-Laird) was implemented [24]. The joint analysis of all combined samples was performed using logistic regression with gender, age and sample cohorts as covariates. Gene annotations were adapted from the University of California at Santa Cruz Genome Browser [25].
Results
Association results of GWAS, replication 1 and combined samples of these two stages
In the replication 1, 49 SNPs were genotyped in 2,202 individuals with SLE and 2,208 healthy controls. After pruning, 45 SNPs that passed quality control were included for final analysis (Table 2 and Additional file 1: Table S1). The association evidence for 15 SNPs with SLE was replicated independently in the replication 1 (P <0.05). When the genotypic data from the GWAS and the replication 1 were combined, we identified two SLE susceptibility genes reaching genome-wide levels of significance (P meta = 5.00 × 10−08): TNFSF4 (rs1418190, odds ratio (OR) = 0.81, P meta = 1.08 × 10−08; rs4916219, OR = 0.80, P meta = 7.77 × 10−09), and IRF8 (rs2934498, OR = 1.25, P meta = 4.97 × 10−9) and another 12 SNPs also showed evidence suggestive of association (Table 2). Our data suggest that the association is independent from our previously reported signals in the neighboring SNPs (rs1234315 and rs2205960) [11], because we found no evidence of strong LD among them (D’ <0.22, r 2 <0.02). To confirm that the two genetic effects detected are independent, we performed logistic regression analysis among them based on the GWAS data, tagged by rs1418190 or rs4916219 and rs1234315 or rs2205960, and showed that each genetic association remained significant (P condition <8.42 × 10−4) after controlling for the effect of each of the remaining two SNPs, using an additive model (Additional file 2: Table S2). In addition, these two novel independent SNPs were confirmed in the haplotype association analysis (Additional file 3: Table S3).
Association results of GWAS, validation stages and combined all samples
We then genotyped these 12 SNPs with suggestive association evidence in the replication 2. The association analysis in replication 2 revealed consistent association with SLE for 7 of 12 SNPs with the GWAS and replication 1 stages. When we combined the genotypic data from the GWAS and two independent replication cohorts, an additional three SNPs within three known SLE susceptibility loci reach genome-wide levels of significance: miR-146a (rs2431697, OR = 0.69, P meta = 1.15 × 10−22), CD44 (rs2732547, OR = 0.82, P meta = 1.55 × 10−11), and TMEM39A (rs12494314, OR = 0.84, P meta = 1.01 × 10−09) (Table 2).
Discussion
In the present study, six SNPs in five known SLE susceptibility loci showed evidence of highly significant association evidence in combined analysis: TNFSF4 (rs1418190, rs4916219), IRF8 (rs2934498), miR-146a (rs2431697), CD44 (rs2732547), and TMEM39A (rs12494314). At 1q25, we identified two new susceptibility SNPs (rs1418190, rs4916219) in TNFSF4, which are independent from two reported SNPs (rs1234315, rs2205960) [11] through logistic regression analysis based on GWAS data. In addition, LD analysis showed that there is no evidence of strong LD among them (D’ <0.22, r 2 <0.02) (Additional file 2: Table S2). TNFSF4 (tumor necrosis factor (ligand) superfamily, member 4) (also known as OX40L), belongs to the TNF ligand family, encodes a cytokine that is involved in T cell antigen-presenting cell interactions [26]. The interaction between OX40L and its receptor (OX40) has a dual effect by delivering a strong co-stimulatory signal to activated effector T-cells and enhances both Th1 and Th2 responses [27], and inhibiting the generation and function of IL-10-producing CD4+ type 1 regulatory T cells [28]. Furthermore, signaling through TNFSF4 is shown to induce B cell activation and differentiation [29], which results in the production of auto-antibodies and immune complexes.
The genetic polymorphisms (rs2280381, rs11644034) in IRF8 (interferon regulatory factor 8) have been confirmed to confer susceptibility to lupus in European populations [30,31]. Although these European SNPs were observed in our GWAS data, no association was discovered with the Chinese population (rs2280381, P = 0.13; rs11644034, P = 0.26). In the present study, we identified a new SNP rs2934498 near rs2280381 (D’ = 0.325, r 2 = 0.008) and rs11644034 (D’ = 0.984, r 2 = 0.049), indicating that IRF8 is also susceptible to SLE in the Chinese population. Previous studies have shown that IRF8 plays a critical role in regulating the differentiation of myeloid and B-cells and can be induced by interferon-γ in macrophages and antigen stimulation within T cells [32,33]. Interestingly, the overexpression of genes induced by type I INF has been widely reported in SLE and other autoimmune diseases [34-36].
As for 5q34, we confirmed an associated variant (rs2431697) between the PTTG1 (pituitary tumor-transforming 1) and the miR-146a (microRNA 146a) genes, which has been identified in previous study in women of European ancestry with SLE. miR-146a is a microRNA (miRNA) that has been shown to be involved in both the regulation of innate and adaptive immune system and tumor progression [37]. Recently, gene expression analysis has revealed that this SNP is not associated with PTTG1 expression levels, but with the miR-146a, where the risk allele correlates with downregulation of the miR-146a, potentially important in SLE etiology [38].
CD44 (CD44 molecule) encodes a transmembrane receptor, which is important for lymphocyte activation, recirculation and homing, apoptosis, hematopoiesis, and tumor metastasis [39,40]. Several studies have identified SPP1, a ligand for CD44, as an SLE risk locus involved in IFN pathways [41,42]. Dysregulation of the IFN system plays critical roles in the pathogenesis of SLE and other closely related autoimmune phenotypes [36]. T cells from SLE patients also display increased and abnormal distribution of CD44 [43], meanwhile, the overexpression of CD44v3 and CD44v6 isoforms in T cells was also observed in the blood of SLE patients, and correlated with disease activity [44]. Several studies have identified multiple variants (rs2732552, rs507230) within or nearby CD44 that are associated with SLE in various populations [45,46]. However, rs2732552 was not covered in our GWAS data and rs507230 did not show any association with SLE in our GWAS data (P = 0.84). In this study, we determined that another SNP rs2732547 near rs507230 (D’ = 0.259, r 2 = 0.067), indicating that CD44 is also associated with SLE in the Chinese population.
At 3q13.33, a coding SNP (rs1132200) in TMEM39A has been reported to be associated with both SLE [31] and multiple sclerosis [47]. However, rs1132200 was not covered in our GWAS data. In the current study, we discovered another SLE-susceptibility SNP (rs12494314) in this region. To date, very few biological data on TMEM39A have been published to provide evidence of its relevance to SLE [31,47].
To investigate the potential molecular mechanism of six significant genetic variants associated with SLE we performed bioinformatics analysis by inferring the relevant biological function from the diverse genomic data and computational prediction. We first utilized ENCODE chromatin state data (three tie 1 cell type: GM12878, K562, H1 human embryonic stem cells) and comprehensive annotations [48] to inspect whether those single nucleotide variants (SNVs) covered by promoter/enhancer/insulator markers (H3K4me1, H3K27ac, p300, CTCF, DNase I hypersensitive site) which indicate the active transcriptional signals. We found rs2431697 (miR-146a), as the leading SNV in GWAS, obtained strong enhancer signals (H3K4me1 and H3K27ac). It is noticeable that rs2431697 (miR-146a) is located in a conserved region by mapping the variant to GERP++ conservation elements [49]. Another leading SNV rs2732547 (CD44) is also marked by p300 and H3K4me1 signals. Therefore, we speculated that those two genetic variants are associated with SLE by affecting the transcription factors binding in their located enhancers. In order to find the affected transcription factors and quantitatively measure the change of binding affinity, we used motifs of different transcription factors from Jasper [50] to scan the surrounding sequences of the above two genetic variants in a different allele state. We then calculated the difference in transcription factor binding score and prioritized them based on the respective P-value. For rs2431697 (miR-146a), we predicted that the binding of transcriptional repressor protein YY1 (YY1 gene) may be disrupted in the corresponding variant region of minor allele G. Importantly, we observed a very significant binding affinity change for androgen receptor (NR3C4) in the rs2732547 (CD44) located region for a different allele. The minor allele A will significantly reduce the binding activity of the androgen receptor and then may result in the development of SLE. The androgen is important in clinical treatment of SLE [51] and the correlation between the androgen receptor and SLE has been investigated by different studies [52,53]. For other associated SNVs, we did not detect sufficient evidence and signals at the transcriptional level.
Conclusions
In conclusion, this study confirmed five previous reported SLE susceptibility loci in Han Chinese population, such as TNFSF4, IRF8, miR-146a, CD44 and TMEM39A. These findings not only provide novel insights into the genetic architecture of SLE but also might highlight the contribution of multiple variants of modest effect.
This study was approved by the ethics committee of The First Affiliated Hospital of Anhui Medical University and conducted according to the principles of the Declaration of Helsinki.
Patient consent was obtained.
Abbreviations
- ACR:
-
American College of Rheumatology
- AHMU:
-
Anhui Medical University
- ANA:
-
antinuclear antibody
- Chr:
-
chromosome
- GWAS:
-
genome-wide association studies
- HWE:
-
Hardy-Weinberg equilibrium
- INF:
-
interferon
- LD:
-
linkage disequilibrium
- MAF:
-
minor allele frequency
- OR:
-
odds ratio
- SLE:
-
systemic lupus erythematosus
- SNP:
-
single nucleotide polymorphism
- SNV:
-
single nucleotide variant
References
Kyogoku C, Tsuchiya N. A compass that points to lupus: genetic studies on type I interferon pathway. Genes Immun. 2007;8:445–55.
Lau CS, Yin G, Mok MY. Ethnic and geographical differences in systemic lupus erythematosus: an overview. Lupus. 2006;15:715–9.
Johnson AE, Gordon C, Palmer RG, Bacon PA. The prevalence and incidence of systemic lupus erythematosus in Birmingham, England. Relationship to ethnicity and country of birth. Arthritis Rheum. 1995;38:551–8.
Seligman VA, Lum RF, Olson JL, Li H, Criswell LA. Demographic differences in the development of lupus nephritis: a retrospective analysis. Am J Med. 2002;112:726–9.
Alarcon GS, Friedman AW, Straaton KV, Moulds JM, Lisse J, Bastian HM, et al. Systemic lupus erythematosus in three ethnic groups: III. A comparison of characteristics early in the natural history of the LUMINA cohort. LUpus in MInority populations: NAture vs. Nurture. Lupus. 1999;8:197–209.
Bongu A, Chang E, Ramsey-Goldman R. Can morbidity and mortality of SLE be improved? Best Pract Res Clin Rheumatol. 2002;16:313–32.
Ward MM, Studenski S. Clinical manifestations of systemic lupus erythematosus. Identification of racial and socioeconomic influences. Arch Intern Med. 1990;150:849–53.
Vyse TJ, Todd JA. Genetic analysis of autoimmune disease. Cell. 1996;85:311–8.
Cui Y, Sheng Y, Zhang X. Genetic susceptibility to SLE: Recent progress from GWAS. J Autoimmun. 2013;41:25–33.
Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–92.
Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–7.
Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Sheng YJ, Gao JP, Li J, Han JW, Xu Q, Hu WL, et al. Follow-up study identifies two novel susceptibility loci PRKCB and 8p11.21 for systemic lupus erythematosus. Rheumatology (Oxford). 2011;50:682–8.
Zhang Z, Cheng Y, Zhou X, Li Y, Gao J, Han J, et al. Polymorphisms at 16p13 are associated with systemic lupus erythematosus in the Chinese population. J Med Genet. 2011;48:69–72.
Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nature genetics. 2009;41:1335–40.
Quan C, Ren YQ, Xiang LH, Sun LD, Xu AE, Gao XH, et al. Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat Genet. 2010;42:614–8.
Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–18.
Sun LD, Xiao FL, Li Y, Zhou WM, Tang HY, Tang XF, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43:690–4.
Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759–63.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58.
Sun LD, Cheng H, Wang ZX, Zhang AP, Wang PG, Xu JH, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–9.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006.
Baum PR, Gayle 3rd RB, Ramsdell F, Srinivasan S, Sorensen RA, Watson ML, et al. Molecular characterization of murine and human OX40/OX40 ligand systems: identification of a human OX40 ligand as the HTLV-1-regulated protein gp34. EMBO J. 1994;13:3992–4001.
Lane P. Role of OX40 signals in coordinating CD4 T cell selection, migration, and cytokine differentiation in T helper (Th)1 and Th2 cells. J Exp Med. 2000;191:201–6.
Ito T, Wang YH, Duramad O, Hanabuchi S, Perng OA, Gilliet M, et al. OX40 ligand shuts down IL-10-producing regulatory T cells. Proc Natl Acad Sci USA. 2006;103:13138–43.
Higgins LM, McDonald SA, Whittle N, Crockett N, Shields JG, MacDonald TT. Regulation of T cell activation in vitro and in vivo by targeting the OX40-OX40 ligand interaction: amelioration of ongoing inflammatory bowel disease with an OX40-IgG fusion protein, but not with an OX40 ligand-IgG fusion protein. J Immunol. 1999;162:486–93.
Cunninghame Graham DS, Morris DL, Bhangale TR, Criswell LA, Syvanen AC, Ronnblom L, et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 2011;7:e1002341.
Lessard CJ, Adrianto I, Ice JA, Wiley GB, Kelly JA, Glenn SB, et al. Identification of IRF8, TMEM39A, and IKZF3-ZPBP2 as susceptibility loci for systemic lupus erythematosus in a large-scale multiracial replication study. Am J Hum Genet. 2012;90:648–60.
Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–84.
Wang H, Morse 3rd HC. IRF8 regulates myeloid and B lymphoid lineage diversification. Immunol Res. 2009;43:109–17.
Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–5.
Baechler EC, Gregersen PK, Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol. 2004;16:801–7.
Baechler EC, Batliwalla FM, Reed AM, Peterson EJ, Gaffney PM, Moser KL, et al. Gene expression profiling in human autoimmunity. Immunol Rev. 2006;210:120–37.
Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–29.
Lofgren SE, Frostegard J, Truedsson L, Pons-Estel BA, D'Alfonso S, Witte T, et al. Genetic association of miRNA-146a with systemic lupus erythematosus in Europeans through decreased expression of the gene. Genes Immun. 2012;13:268–74.
Vivers S, Dransfield I, Hart SP. Role of macrophage CD44 in the disposal of inflammatory cell corpses. Clin Sci (Lond). 2002;103:441–9.
Ponta H, Sherman L, Herrlich PA. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol. 2003;4:33–45.
Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–9.
Kariuki SN, Moore JG, Kirou KA, Crow MK, Utset TO, Niewold TB. Age- and gender-specific modulation of serum osteopontin and interferon-alpha by osteopontin genotype in systemic lupus erythematosus. Genes Immun. 2009;10:487–94.
Crispin JC, Tsokos GC. Novel molecular targets in the treatment of systemic lupus erythematosus. Autoimmun Rev. 2008;7:256–61.
Crispin JC, Keenan BT, Finnell MD, Bermas BL, Schur P, Massarotti E, et al. Expression of CD44 variant isoforms CD44v3 and CD44v6 is increased on T cells from patients with systemic lupus erythematosus and is correlated with disease activity. Arthritis Rheum. 2010;62:1431–7.
Ramos PS, Williams AH, Ziegler JT, Comeau ME, Guy RT, Lessard CJ, et al. Genetic analyses of interferon pathway-related genes reveal multiple new loci associated with systemic lupus erythematosus. Arthritis Rheum. 2011;63:2049–57.
Lessard CJ, Adrianto I, Kelly JA, Kaufman KM, Grundahl KM, Adler A, et al. Identification of a systemic lupus erythematosus susceptibility locus at 11p13 between PDHX and CD44 in a multiethnic study. Am J Hum Genet. 2011;88:83–91.
Comprehensive follow-up of the first genome-wide association study of multiple sclerosis identifies KIF21B and TMEM39A as susceptibility loci. Hum Mol Genet. 2010;19:953–62.
Li MJ, Sham PC, Wang J. Genetic variant representation, annotation and prioritization in the post-GWAS era. Cell Res. 2012;22:1505–8.
Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13.
Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–4.
Meier CR, Sturkenboom MC, Cohen AS, Jick H. Postmenopausal estrogen replacement therapy and the risk of developing systemic lupus erythematosus or discoid lupus. J Rheumatol. 1998;25:1515–9.
Tessnow AH, Olsen NJ, Kovacs WJ. Expression of humoral autoimmunity is related to androgen receptor CAG repeat length in men with systemic lupus erythematosus. J Clin Immunol. 2011;31:567–73.
Robeva R, Tanev D, Andonova S, Kirilov G, Savov A, Stoycheva M, et al. Androgen receptor (CAG)n polymorphism and androgen levels in women with systemic lupus erythematosus and healthy controls. Rheumatol Int. 2013;33:2031–8.
Acknowledgements
We thank all study participants and all the volunteers who have so willingly participated in this study, thus making this study possible. This work was supported by grants from National Key Basic Research Program of China (2014CB541901, 2012CB722404 and 2011CB512103), the National Natural Science Foundation of China (81402590, 81371722, 81320108016 and 81171505), the Research Project of Chinese Ministry of Education (No.213018A), the Program for New Century Excellent Talents in University (NCET-12-0600), the Natural Science Fund of Anhui province (1408085MKL27) and Youth Science Training Program of National Natural Science Foundation of China (2010kj10).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XJZ, YCu and YJS conceived of this study, obtained financial support, participated in the study design and revised the manuscript. YJS, JHX and YGW were responsible for sample selection, genotyping and project management and drafted the manuscript. YL, LLW, CY, YCh, LL, ZWZ, YYC, LLY, LDS and SY conducted sample selection and data management, undertook recruitment, collected phenotyping data, undertook related data handling and calculation, managed recruitment, obtained biological samples and helped to revise the manuscript. FSZ, JPG and XFT performed genotyping analysis and helped to revise the manuscript. XYY, HYT, XDZ and XBZ performed data processing and statistical analysis and revised the manuscript. All authors read and approved the final manuscript.
Yu-jun Sheng, Jian-hua Xu and Yong-gui Wu contributed equally to this work.
Additional files
Additional file 1: Table S1.
Summary of the association results for 34 SNPs in the GWAS and replication 1 and combined studies.
Additional file 2: Table S2.
Conditional analysis between two new susceptibility SNPs (rs1418190, rs4916219) and two reported SNPs (rs1234315, rs2205960) in GWAS samples.
Additional file 3: Table S3.
Haplotypic association for markers in TNFSF4 locus with the risk of SLE.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Sheng, Yj., Xu, Jh., Wu, Yg. et al. Association analyses confirm five susceptibility loci for systemic lupus erythematosus in the Han Chinese population. Arthritis Res Ther 17, 85 (2015). https://doi.org/10.1186/s13075-015-0602-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13075-015-0602-9