Abstract
Background
Differences in the expression of variants across ethnic groups in the systemic lupus erythematosus (SLE) patients have been well documented. However, the genetic architecture in the Thai population has not been thoroughly examined. In this study, we carried out genome-wide association study (GWAS) in the Thai population.
Methods
Two GWAS cohorts were independently collected and genotyped: discovery dataset (487 SLE cases and 1606 healthy controls) and replication dataset (405 SLE cases and 1590 unrelated disease controls). Data were imputed to the density of the 1000 Genomes Project Phase 3. Association studies were performed based on different genetic models, and pathway enrichment analysis was further examined. In addition, the performance of disease risk estimation for individuals in Thai GWAS was assessed based on the polygenic risk score (PRS) model trained by other Asian populations.
Results
Previous findings on SLE susceptible alleles were well replicated in the two GWAS. The SNPs on HLA class II (rs9270970, A>G, OR = 1.82, p value = 3.61E−26), STAT4 (rs7582694, C>G, OR = 1.57, p value = 8.21E−16), GTF2I (rs73366469, A>G, OR = 1.73, p value = 2.42E−11), and FAM167A-BLK allele (rs13277113, A>G, OR = 0.68, p value = 1.58E−09) were significantly associated with SLE in Thai population. Meta-analysis of the two GWAS identified a novel locus at the FBN2 that was specifically associated with SLE in the Thai population (rs74989671, A>G, OR = 1.54, p value = 1.61E−08). Functional analysis showed that rs74989671 resided in a peak of H3K36me3 derived from CD14+ monocytes and H3K4me1 from T lymphocytes. In addition, we showed that the PRS model trained from the Chinese population could be applied in individuals of Thai ancestry, with the area under the receiver-operator curve (AUC) achieving 0.76 for this predictor.
Conclusions
We demonstrated the genetic architecture of SLE in the Thai population and identified a novel locus associated with SLE. Also, our study suggested a potential use of the PRS model from the Chinese population to estimate the disease risk for individuals of Thai ancestry.
Similar content being viewed by others
Background
The systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by loss of tolerance to self-antigens, inappropriate immune activation, and inflammation [1]. The severity is various depending on affected organs [2]. Genetic susceptibility has been widely accepted as one of the critical factors driving disease development [2]. Recently, the genetic architecture of SLE has been examined worldwide [3]. Using GWAS, more than 90 loci have been found associated with SLE across at least four ethnic groups, including Han Chinese, European, North America, and Africa [4, 5]. The strongest signal was identified at the HLA class II allele, which replicated in all of the different populations [4]. These findings indicate critical biological mechanisms underlying the disease, which will be the candidate in further functional studies [6].
However, heterogeneity of disease between different ethnicities drives a question of whether genetic background in different ancestries could affect the clinical manifestations. It is known that Asian SLE patients have higher disease severity compared to Europeans [2]. However, only a few studies on SLE associations that were based on candidate genes were performed in the Thai population [7,8,9,10,11]. In this study, we conducted GWAS using the SLE samples collected from two tertiary referral hospitals in Thailand. We aim to replicate known SLE-associated variants in the Thai population and identify novel SNPs associated with SLE.
Methods
Sample collection and preparation
We calculated the power of our study by using an online tool called Genetic association study (GAS) Power Calculator [12]. With 800 cases and 1600 controls at 5E−08 significant level, we obtained 0.934 expected power for the study. The EDTA blood samples from SLE patients (n = 487) were collected at King Chulalongkorn Memorial Hospital as cases for the discovery phase. All procedures were approved by the ethical committee from the Faculty of Medicine, Chulalongkorn University (COA no. 923/2017). For the replication cohort, the samples (n = 405) were collected from the Rheumatology clinic, Ramathibodi Hospital, with ethical approval from the Faculty of Medicine, Mahidol University (MURA no. 2015/731, EC no. 590223, Protocol-ID 12-58-12). All patients were carefully recruited regarding the criteria from the American College of Rheumatology (ACR) [13]. Patients’ demographic data from both datasets have been summarized in Table 1. For healthy controls (n = 1606) and unrelated disease controls including breast cancer, periodontitis, tuberculosis, drug-induced liver injury, epileptic encephalopathy, dengue hemorrhagic fever, thalassemia, and cardiomyopathy (n = 1590), data were provided from the Department of Medical Science, Ministry of Public Health, Thailand.
DNA extraction
Buffy coats were extracted using the QIAGEN® EZ1® DNA blood kit (QIAGEN GmbH, Hilden, Germany). We used 200 μl of a buffy coat as recommended by the manufacturer’s instruction. Buffy coat samples were transferred into tube or sample cartridge for EZ1 Advanced XL (QIAGEN GmbH, Hilden, Germany) and extracted using EZ1® Advanced XL DNA Buffy coat protocol. From this protocol, DNA was eluted at 200 μl. DNA was diluted and quantitated using Qubit™ dsDNA BR Assay Kit according to the manufacturing protocol (Invitrogen, Thermo Fisher Scientific, MA, USA).
Genotyping and quality control
Genotyping was performed using Infinium Asian Screening Array-24 v1.0 BeadChip with 659,184 SNPs (Illumina, San Diego, CA, USA) at the Department of Medical Sciences (DMSC, Ministry of Public Health, Thailand) based on the protocol recommended by the manufacturer. The Genome Studio data analysis software v2011.1 (Illumina, San Diego, CA, USA) was used for calling genotypes. Samples and SNP markers were tested for quality control (QC) using PLINK genomic analysis software (v1.90b5.4) [14]. Individuals with autosomal genotype call rate ≤ 0.98, gender inconsistency based on heterozygosity rate of X chromosome (maleTh = 0.8, femaleTh = 0.2), and genome-wide estimates of identity-by-descent (pihat) ≥ 0.185 (3rd generation) were excluded from analysis. SNPs with more than 5% missing genotyping rate or significant deviation of Hardy-Weinberg equilibrium (p value ≤ 1 × 10−8) were also removed. After quality control (QC), we obtained a dataset of 2041 individuals with 421,909 variants for the discovery phase and 1955 individuals with 446,139 variants for replication. The flow diagram of the analysis process is shown in Fig. 1a.
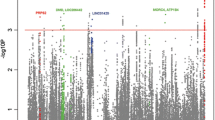
Quality control and dataset preparation flow diagram of both discovery and validation datasets. The flow diagram was modified from the PRISMA flow diagram [15] (a). Manhattan plot on the meta-analysis result of the two SLE GWAS datasets in the Thai population using R-Bioconductor package qqman (b)
GWAS data imputation
Pre-phasing was performed using SHAPEIT [16]. After that, genotype data for individuals was imputed to the density of the 1000 Genomes Project Phase 3 reference using IMPUTE2 [17]. After all the QC processing, 6,657,806 were left for further analysis. The processed data were publicly available at http://2anp.2.vu/GWAS_SLE_Thailand.
Association study, meta-analysis, and statistical analysis
The association studies were conducted by using SNPTEST [18], and the factored spectrally transformed linear mixed models (FaST-LMM v.0.2.32) program [19]. The results from FaST-LMM were analyzed and visualized by RStudio to obtain genomic inflation factor (λ), quantile-quantile plot, and Manhattan plot [20]. The SNPs with p value ≤ 1 × 10−5 were plotted to obtain the regional plot by using LocusZoom [21]. Haplotype block and linkage disequilibrium structure were analyzed by Haploview software version 4.2 [22]. The characterized SLE susceptible loci were downloaded from a previous study [23] and GWAS catalogue (the NHGRI-EBI catalogue of published genome-wide association studies). Meta-analysis was studied based on the inverse variant strategy in the METAL program [24]. The genetic inheritance pattern was calculated from the frequency of different genotyping on risk alleles using R-Bioconductor. Simultaneously, functional annotation was predicted by using SNPnexus, which applied data from the Reactome database [25]. The histone markers and transcription factor binding sites were predicted from an online tool called HaploReg V4.1 [26].
Polygenic risk score calculation
Lassosum [27] was used to calculate PRS for individuals. The summary statistics for SLE association in East Asians [28], involving 2618 cases and 7446 controls with Chinese ancestry, were used to train the model. The area under the ROC curve (AUC) was calculated using R package pROC [29].
Results
Known SLE associations found in the Thai population
In the discovery dataset, the association studies were initially performed using healthy controls (n = 1606) and SLE patients (n = 487) collected from King Chulalongkorn Memorial Hospital. Regarding the result, we found that variants at the HLA class II regions were strongly associated with SLE (p value < 5E−08). Similarly, GWAS from 405 SLE cases and 1590 non-immune-mediated disease controls found variants at the HLA class II regions reached the genome-wide significant threshold (p value < 5E−08). Our findings were consistent with previous reports in other ethnic groups [30]. Inflation factors from both datasets were calculated as reported in Supplementary figure 1.
Subsequently, a meta-analysis of the two Thai GWAS was carried out, and we systematically examined associations across the 90 known SLE-associated loci, which were collected from the GWAS catalogue (https://www.ebi.ac.uk/gwas/) and previous review articles [23]. Of these loci, the HLA-DQA1, HLA-DRB1, STAT4, FAM167A-BLK, and GTF2I loci have reached the genome-wide significant threshold (p value < 5E−08; Fig. 1b, Table 2) in Thai population, and the variants at the PROS1C1, NOTCH4, HCP5, C6orf10, TAP2, TNFSF4, RasGRP3, TERT, TNPO3-IRF5, CXCR5, GPR19, SLC15A4, and ITGAM loci showed suggestive evidence of associations with SLE (p value < 5E−05, Supplementary Table 1). These loci have been found in several ancestries, including Han Chinese, Korean, North American, European, African, and Hispanic populations [31, 32].
We noticed that some of the previously characterized nonsynonymous polymorphisms also showed certain evidence of association (p value < 0.05) in Thai population, such as rs11235604 (ATG16L2, R58W), rs13306575 (NCF2, R395W), rs1990760 (IFIH1, A946T), rs3734266 (UHRF1BP1, Q454L), rs2841280 (PLD4, E27Q), and rs2230926 (TNFAIP3, F127S). Details of these associations are summarized in Table 3. All significant variants were calculated for Hardy-Weinberg equilibrium, as reported in Supplementary Table 2.
Identification of novel loci associated with SLE
Excluding the variants at the known SLE-associated loci, we discovered a novel variant on FNB2 (rs74989671, OR = 1.54, p value = 1.61E−08) specifically associated with SLE in Thai population (Figs. 1b and 2a, Table 2) when comparing the association in Europeans (OR = 0.998, p value = 0.979) and in Chinese populations (OR = 0.982, p value = 0.692) [28]. Further analyses based on different genetic inheritance models suggested that the disease risk was associated with the copy number of risk alleles that the individuals carried (additive model) (Table 4). Three SNPs on FBN2 loci (rs74989671, rs35983844, rs6595836) showed linkage disequilibrium (LD r2 = 0.82) (Fig. 2b, Supplementary Table 1). Of these variants, rs74989671 was found to locate within the peak of H3K36me3 derived from CD14-positive monocytes and H3K4me1 (associated with active enhancers) derived from the primary T cells (Fig. 2c).
Regional plot of novel SLE susceptible variants on FBN2 locus with their relative variants around FBN2 locus (a). Haplotype block of significant variants on FBN2 locus with their correlation to show linkage disequilibrium between SNPs (b). The picture illustrated histone markers overlapped with FBN2 SNP site (c)
In addition, we found variants at the ATP6V1B1, MIR4472-2, MYO5C, ADCY5, and DGKG, showing suggestive evidence of associations with SLE in Thai population (p value < 5E−05) (Supplementary Figure 2, Supplementary Table 1). Though these polymorphisms are likely to associate with Thai SLE patients, an independent GWAS dataset of SLE patients with Thai background is needed for further validation.
In silico functional annotation of SLE-associated variants in Thai population
To understand the biological meaning underlying the SLE-associated loci in the Thai population, we performed the pathway analysis using the SNPnexus program [25]. Variants with p value < 5E−05 were involved in this study. Notably, we found that 50% of all variants were located within the coding region, by which 10% is nonsynonymous polymorphisms. Pathway analysis results revealed that SLE-associated variants were highly enriched in the regulation of interferon signaling, PD-1 signaling, MHC-class II antigen presentation, TCR/BCR signaling, cytokine signaling, TNF signaling, NOTCH4 signaling, calcium-activated potassium channels, and cell-cell junction organization pathways. Furthermore, we found that extracellular matrix organization was significant in our results (Fig. 3). It indicated that Thai SLE patients might have a higher risk of fibrosis-associated inflammation.
Polygenic risk score prediction for the individuals
To apply the GWAS result to predict the Thai SLE outcome, we also tested the hypothesis of whether the PRS models trained by individuals with Chinese ancestry could be applied for Thai SLE patients. We calculated PRS for individuals in the Thai GWAS, based on the training data from the Chinese population (2618 cases and 7446 controls) [28]. Significantly, the PRS for SLE cases were higher than controls (mean difference = 0.89; p value = 2.2E−16; Fig. 4a), and the area under the receiver-operator curve (AUC) achieved 0.76 for this predictor. This analysis indicated the potential application for the PRS in the Thai population, based on the results from other Asian populations. Regarding the analysis, this might be a clue for predicting an outcome of SLE clinical characteristics in Thai SLE patients, and it is a good source for further genetic analysis to identify actual SLE pathogenesis in the different ancestry.
The graph shows the polygenic risk score calculation and the mean difference between SLE and healthy controls (a). The circular plot showed loci which identified in this study at individual chromosomes using package Rcircos [33] (b)
Discussion
The present study is the first largest GWAS cohort conducted among Thai SLE patients. The highest significant association in the region of HLA class II was consistent with previous reports from other ethnic groups [30]. Since there are vast differences of HLA class II allele frequency among populations and sophisticated genetic structure, the study of the specific HLA class II haplotype is needed. Currently, there were two publications reported about specific HLA haplotypes in Thai SLE patients. First, the fine genetic mapping of the HLA allele from SLE patients in the northern part of Thailand has identified the association of HLA-DRB5*01:01 and HLA-DRB1*16:02 [34]. Secondly, HLA haplotype analysis found HLA-DRB1*15:02 and HLA-DQ*05:01 associated with Thai SLE patients [35]. Further study on the HLA class II allele using whole-genome sequencing or exome sequencing might be helpful to specify the impact of the HLA class II allele on Thai SLE patients.
Apart from the HLA class II alleles, our study found variants in STAT4, GTF2I, and BLK regions. For BLK locus, this gene encoded for Src-tyrosine kinase, which is an important signaling molecule under B cell development [36]. This gene showed protein-protein interaction with BANK1 (B cell-specific cytoplasmic protein involved in B cell receptor signaling) and might plausibly involve in dysregulation of the B cell receptor, which is a common feature found in SLE patients [37]. For STAT4 and GTF2I alleles, these genes are encoded for the transcription factors that mediate many immune-related genes and inflammatory cytokine transcription machinery. Both BLK and STAT4 loci have been reported as SLE susceptible alleles in Thai SLE patients recently [7], whereas GTF2I locus has firstly identified in our study. Interestingly, the variants on STAT4 and GTF2I loci were correlated with lupus nephritis (LN) in the various SLE ancestries [32]. The GTF2I allele was likely to be specific in Asian background, mainly Han Chinese [38].
Our analysis found several LN-susceptible loci such as IRF5 [39, 40], ITGAM [9, 41], IKZF1 [42], and TNFSF4 [43]. While IKZF1 is a co-transcription factor with STAT-4 mediated Th1 lymphocyte differentiation and interferon pathways [44], the TNFSF4 locus, also called OX40L, encoded for the TNF superfamily ligand, which actively stimulates CD4+ T cell activation [43]. Study in the Finnish and Swedish SLE patients found the correlation of ITGAM with cutaneous discoid lupus erythematosus (DLE) and LN as well as Ro/SSA auto-antibody positive [45]. Not only LN, but we also found several loci that have been verified in the specific sub-phenotype of SLE patients. For example, our result found a variant on ETS1, which previously showed association with juvenile SLE, as well as a variant on RasGRP3, which was involved in malar rash or discoid rash [42]. The recent SLE susceptible loci identified in the cardiac manifestation of neonatal lupus, NOTCH4, was found in our results [46].
Note that we found some of the known SNPs which are nonsynonymous variants such as NCF2 [47], IFIH1 [48], TNFAIP3 [49], UHRF1BP1 [50], ATG16L2 [51], and PLD4 [52]. A few pieces of evidence have revealed the impact of those variants on various pathways including neutrophil extracellular traps (NETs) formation [53], sensor molecule to detect viral genome inside cells [54], a negative regulator for NF-kB transcription factors [55], and a negative regulator of cell growth [56]. These pathways resembled with our functional enrichment pathways analysis. Interestingly, our results found extracellular matrix organization (ECM) pathways associated with Thai SLE patients. Previously, single-cell transcriptome analysis in non-responder LN patients highlighted the upregulated genes in the ECM pathway correlated with treatment failure [57]. The ECM reflected the active fibrotic process, which was a marker of poor prognosis LN [58]. Remarkably, the prevalence of severe LN was high in the South East Asian ethnic included Thai [59]. As regards our SLE patients’ demographic data, we found that the frequency of clinical phenotype was roughly similar to other ethnic [60]. The LN has the highest abundance found among Thai SLE patients. Thus, our results supported that genetic background was a pivotal factor driving a severe LN among Thai SLE patients. Taken together, these pieces of evidence could justify the link between genetic variants and clinical involvement in Thai SLE patients.
The study of known SNPs showed most of the polymorphisms resembled with previous reports in Thais, such as ARID5B, TNFSF4, BANK1, TNFAIP3, CXCR5 SLC15A, ITGAM, WDFY4, ETS1, and BLK [7,8,9,10,11]. It confirmed that our analysis processes were reliable. Noticeably, the allele frequency of ITGAM was higher among Thai SLE when compared to Chinese Hong Kong [9], but has no association with Japanese and Korean background [61]. Thus, this implies the specificity of these variants to the Thai SLE patients. Although we did not recognize polymorphisms on chromosome 11q23.3 (rs11603023 on PHLDB1 and rs638893 on DDX6), which has been identified in the Thais’ SLE, our meta-analysis enhanced signal from rs10845606 on GPR19 allele which does not correlate with Thai SLE patients previously [8].
It is noteworthy that meta-analysis in the Thai population discovered novel SLE susceptible variants on FBN2. The FBN2 allele is located on a chromosome 5 encoded protein called fibrillin-2 [62]. Fibrillins-2 is one of the glycoprotein components incorporated extracellularly on microfibrils and is essential in bone, muscle, and extracellular matrix formation [63]. It is well known that mutation of FBN2 leads to dominant heritable connective tissue disorders [64]. Importantly, a recent review article has gained insight on fibrillin-2 as a critical mediator that binds to transforming growth factor-beta (TGF-β) during extracellular matrix formation [65]. The TLR9/TGF-β1/PDGF-β pathway was excessively activated in peripheral mononuclear cells isolated from LN patients [66]. Besides, the upregulation of FBN2 correlated with fibrosis prevalence in the spontaneous LN developed mouse model (SWR X NZB1 F1) [67]. Although the function of FBN2 in SLE is unclear, collective evidence led us to hypothesize that this variant might drive either fibrosis-associated inflammation or inflammatory induction during disease pathogenesis. Due to whole-genome sequencing data in the Thai population is lacking, further study using FBN2 target sequencing, whole-genome sequencing, and variant functional characterization in a large cohort is needed. This knowledge could be useful to identify rare coding variants and genetic propensity eliciting SLE pathogenesis in Thais.
Note that some of the variants were previously characterized in other autoimmune diseases, including rheumatoid arthritis and primary Sjögren syndrome (pSS). It, therefore, indicates the sharing of underlying genetic factors between autoimmune disease. However, predisposing factors which could affect clinical manifestation driving different autoimmune disease outcome has not been elucidated yet. Recently, the GRS (genetic risk score) has been widely adopted to predict disease outcomes from genetic variants [68]. The previous studies in SLE showed that overall mortality was higher in the striking GRS SLE patients; also, the high cumulative genetic risk could predict the specific organ damages such as proliferative nephritis and cardiovascular disease [69]. Our study showed a high sensitivity for using polygenic risk scored as a marker for SLE disease development in the Thai population. It is exciting for further study to calculate the genetic risk score and specific clinical manifestation among Thai SLE patients.
Conclusions
In conclusion, our study reported susceptible loci of SLE patients in Thai ancestry, which were variants on the HLA class II allele, STAT4, GTF2I, and BLK. Additionally, we confirmed those variants which had been reported previously in the Thai populations, which were ARID5B, TNFSF4, BANK1, TNFAIP3, CXCR5 SLC15A, ITGAM, WDFY4, and ETS1. Interestingly, we identified novel variants associated with the Thai SLE patients, which were on the FNB2 allele. Summary loci associated with the Thai SLE were seen in Fig. 4b. Functional annotation analysis highlighted extracellular matrix organization pathways specific to the Thai population. The PRS using GWAS data is useful for SLE prediction with sensitivity and specificity of more than 70%. Further whole-genome sequencing study with a large sample size might help to validate our results. Finally, our finding provides the necessary genetic background susceptible to SLE disease, expanding the number of molecular targets for treatment options.
Availability of data and materials
The complete results from the two Thai GWAS datasets and the GWAS meta-analysis are publicly available for download at http://2anp.2.vu/GWAS_SLE_Thailand.
Abbreviations
- GWAS:
-
Genome-wide association study
- SLE:
-
Systemic lupus erythematosus
- SNP:
-
Single nucleotide polymorphism
- HLA:
-
Human leukocyte antigen
- MHC:
-
Major histocompatibility
- CHR:
-
Chromosome
- MAF:
-
Minor allele frequency
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- bp:
-
Base pair
- BCR:
-
B cell receptor
- TCR:
-
T cell receptor
- IL:
-
Interleukin
- PD:
-
Program cell death
- MDA:
-
Melanoma differentiation-associated gene 5
References
Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358(9):929–39.
Ceccarelli F, Perricone C, Borgiani P, Ciccacci C, Rufini S, Cipriano E, Alessandri C, Spinelli FR, Sili Scavalli A, Novelli G, et al. Genetic factors in systemic lupus erythematosus: contribution to disease phenotype. J Immunol Res. 2015;2015:745647.
Goulielmos GN, Zervou MI, Vazgiourakis VM, Ghodke-Puranik Y, Garyfallos A, Niewold TB. The genetics and molecular pathogenesis of systemic lupus erythematosus (SLE) in populations of different ancestry. Gene. 2018;668:59–72.
Kwon YC, Chun S, Kim K, Mak A. Update on the genetics of systemic lupus erythematosus: genome-wide association studies and beyond. Cells. 2019;8(10):1180–96.
Julia A, Lopez-Longo FJ, Perez Venegas JJ, Bonas-Guarch S, Olive A, Andreu JL, Aguirre-Zamorano MA, Vela P, Nolla JM, de la Fuente JLM, et al. Genome-wide association study meta-analysis identifies five new loci for systemic lupus erythematosus. Arthritis Res Ther. 2018;20(1):100.
Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep. 2012;39(12):10627–35.
Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, Hirankarn N, Ying D, Pan HF, Mok CC, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6(2):e1000841.
Yang W, Tang H, Zhang Y, Tang X, Zhang J, Sun L, Yang J, Cui Y, Zhang L, Hirankarn N, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am J Hum Genet. 2013;92(1):41–51.
Yang W, Zhao M, Hirankarn N, Lau CS, Mok CC, Chan TM, Wong RW, Lee KW, Mok MY, Wong SN, et al. ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Hum Mol Genet. 2009;18(11):2063–70.
Zhang H, Zhang Y, Wang YF, Morris D, Hirankarn N, Sheng Y, Shen J, Pan HF, Yang J, Yang S, et al. Meta-analysis of GWAS on both Chinese and European populations identifies GPR173 as a novel X chromosome susceptibility gene for SLE. Arthritis Res Ther. 2018;20(1):92.
Zhang J, Zhang Y, Yang J, Zhang L, Sun L, Pan HF, Hirankarn N, Ying D, Zeng S, Lee TL, et al. Three SNPs in chromosome 11q23.3 are independently associated with systemic lupus erythematosus in Asians. Hum Mol Genet. 2014;23(2):524–33.
Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–13.
Aringer M, Costenbader K, Daikh D, Brinks R, Mosca M, Ramsey-Goldman R, Smolen JS, Wofsy D, Boumpas DT, Kamen DL, et al. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann Rheum Dis. 2019;78(9):1151–9.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
Delaneau O, Marchini J, Genomes Project C, Genomes Project C. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014;5:3934.
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–9.
Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511.
Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8(10):833–5.
Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv; 2014. https://doi.org/10.1101/005165.
Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5.
Chen L, Morris DL, Vyse TJ. Genetic advances in systemic lupus erythematosus: an update. Curr Opin Rheumatol. 2017;29(5):423–33.
Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1.
Dayem Ullah AZ, Oscanoa J, Wang J, Nagano A, Lemoine NR, Chelala C. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res. 2018;46(W1):W109–13.
Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–81.
Mak TSH, Porsch RM, Choi SW, Zhou X, Sham PC. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol. 2017;41(6):469–80.
Wang Y-F, Zhang Y, Lin Z, Zhang H, Wang T-Y, Cao Y, Morris DL, Sheng Y, Yin X, Zhong S-L et al: Identification of 38 novel loci for systemic lupus erythematosus and genetic heterogeneity that may underly population disparities in this disease. 2020:2020.2004.2012.037622.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77.
Armstrong DL, Zidovetzki R, Alarcon-Riquelme ME, Tsao BP, Criswell LA, Kimberly RP, Harley JB, Sivils KL, Vyse TJ, Gaffney PM, et al. GWAS identifies novel SLE susceptibility genes and explains the association of the HLA region. Genes Immun. 2014;15(6):347–54.
Meng Y, He Y, Zhang J, Xie Q, Yang M, Chen Y, Wu Y. Association of GTF2I gene polymorphisms with renal involvement of systemic lupus erythematosus in a Chinese population. Medicine (Baltimore). 2019;98(31):e16716.
Oparina N, Martinez-Bueno M, Alarcon-Riquelme ME. An update on the genetics of systemic lupus erythematosus. Curr Opin Rheumatol. 2019;31(6):659–68.
Zhang H, Meltzer P, Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14:244.
Louthrenoo W, Kasitanon N, Wichainun R, Wangkaew S, Sukitawut W, Ohnogi Y, Hong GH, Kuwata S, Takeuchi F. The genetic contribution of HLA-DRB5*01:01 to systemic lupus erythematosus in Thailand. Int J Immunogenet. 2013;40(2):126–30.
Sirikong M, Tsuchiya N, Chandanayingyong D, Bejrachandra S, Suthipinittharm P, Luangtrakool K, Srinak D, Thongpradit R, Siriboonrit U, Tokunaga K. Association of HLA-DRB1*1502-DQB1*0501 haplotype with susceptibility to systemic lupus erythematosus in Thais. Tissue Antigens. 2002;59(2):113–7.
Castillejo-Lopez C, Delgado-Vega AM, Wojcik J, Kozyrev SV, Thavathiru E, Wu YY, Sanchez E, Pollmann D, Lopez-Egido JR, Fineschi S, et al. Genetic and physical interaction of the B-cell systemic lupus erythematosus-associated genes BANK1 and BLK. Ann Rheum Dis. 2012;71(1):136–42.
Ramirez-Bello J, Jimenez-Morales S, Montufar-Robles I, Fragoso JM, Barbosa-Cobos RE, Saavedra MA, Sanchez-Munoz F. BLK and BANK1 polymorphisms and interactions are associated in Mexican patients with systemic lupus erythematosus. Inflamm Res. 2019;68(8):705–13.
Li Y, Li P, Chen S, Wu Z, Li J, Zhang S, Cao C, Wang L, Liu B, Zhang F, et al. Association of GTF2I and GTF2IRD1 polymorphisms with systemic lupus erythematosus in a Chinese Han population. Clin Exp Rheumatol. 2015;33(5):632–8.
Qin L, Lv J, Zhou X, Hou P, Yang H, Zhang H. Association of IRF5 gene polymorphisms and lupus nephritis in a Chinese population. Nephrology (Carlton). 2010;15(7):710–3.
Watkins AA, Yasuda K, Wilson GE, Aprahamian T, Xie Y, Maganto-Garcia E, Shukla P, Oberlander L, Laskow B, Menn-Josephy H, et al. IRF5 deficiency ameliorates lupus but promotes atherosclerosis and metabolic dysfunction in a mouse model of lupus-associated atherosclerosis. J Immunol. 2015;194(4):1467–79.
Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358(9):900–9.
He CF, Liu YS, Cheng YL, Gao JP, Pan TM, Han JW, Quan C, Sun LD, Zheng HF, Zuo XB, et al. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus. 2010;19(10):1181–6.
Farres MN, Al-Zifzaf DS, Aly AA, Abd Raboh NM. OX40/OX40L in systemic lupus erythematosus: association with disease activity and lupus nephritis. Ann Saudi Med. 2011;31(1):29–34.
Hu SJ, Wen LL, Hu X, Yin XY, Cui Y, Yang S, Zhang XJ. IKZF1: a critical role in the pathogenesis of systemic lupus erythematosus? Mod Rheumatol. 2013;23(2):205–9.
Jarvinen TM, Hellquist A, Koskenmies S, Einarsdottir E, Panelius J, Hasan T, Julkunen H, Padyukov L, Kvarnstrom M, Wahren-Herlenius M, et al. Polymorphisms of the ITGAM gene confer higher risk of discoid cutaneous than of systemic lupus erythematosus. PLoS One. 2010;5(12):e14212.
Clancy RM, Marion MC, Kaufman KM, Ramos PS, Adler A, International Consortium on Systemic Lupus Erythematosus G, Harley JB, Langefeld CD, Buyon JP. Identification of candidate loci at 6p21 and 21q22 in a genome-wide association study of cardiac manifestations of neonatal lupus. Arthritis Rheum. 2010;62(11):3415–24.
Kim-Howard X, Sun C, Molineros JE, Maiti AK, Chandru H, Adler A, Wiley GB, Kaufman KM, Kottyan L, Guthridge JM, et al. Allelic heterogeneity in NCF2 associated with systemic lupus erythematosus (SLE) susceptibility across four ethnic populations. Hum Mol Genet. 2014;23(6):1656–68.
Zhang J, Liu X, Meng Y, Wu H, Wu Y, Yang B, Wang L. Autoimmune disease associated IFIH1 single nucleotide polymorphism related with IL-18 serum levels in Chinese systemic lupus erythematosus patients. Sci Rep. 2018;8(1):9442.
Liu X, Qin H, Wu J, Xu J. Association of TNFAIP3 and TNIP1 polymorphisms with systemic lupus erythematosus risk: a meta-analysis. Gene. 2018;668:155–65.
Zhang Y, Yang W, Mok CC, Chan TM, Wong RW, Mok MY, Lee KW, Wong SN, Leung AM, Lee TL, et al. Two missense variants in UHRF1BP1 are independently associated with systemic lupus erythematosus in Hong Kong Chinese. Genes Immun. 2011;12(3):231–4.
Ishibashi K, Fujita N, Kanno E, Omori H, Yoshimori T, Itoh T, Fukuda M. Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12-5-16L2 complex. Autophagy. 2011;7(12):1500–13.
Akizuki S, Ishigaki K, Kochi Y, Law SM, Matsuo K, Ohmura K, Suzuki A, Nakayama M, Iizuka Y, Koseki H, et al. PLD4 is a genetic determinant to systemic lupus erythematosus and involved in murine autoimmune phenotypes. Ann Rheum Dis. 2019;78(4):509–18.
Jacob CO, Eisenstein M, Dinauer MC, Ming W, Liu Q, John S, Quismorio FP Jr, Reiff A, Myones BL, Kaufman KM, et al. Lupus-associated causal mutation in neutrophil cytosolic factor 2 (NCF2) brings unique insights to the structure and function of NADPH oxidase. Proc Natl Acad Sci U S A. 2012;109(2):E59–67.
Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, Mikolaitis RA, Guerrero G, Utset TO, Drevlow BE, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-alpha and serologic autoimmunity in lupus patients. J Immunol. 2011;187(3):1298–303.
Das T, Chen Z, Hendriks RW, Kool M. A20/tumor necrosis factor alpha-induced protein 3 in immune cells controls development of autoinflammation and autoimmunity: lessons from mouse models. Front Immunol. 2018;9:104.
Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene. 2004;23(46):7601–10.
Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabothu S, Izmirly P, Clancy R, Belmont HM, Koenigsberg M, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol. 2019;20(7):915–27.
Hsieh C, Chang A, Brandt D, Guttikonda R, Utset TO, Clark MR. Predicting outcomes of lupus nephritis with tubulointerstitial inflammation and scarring. Arthritis Care Res (Hoboken). 2011;63(6):865–74.
Avihingsanon Y, Hirankarn N. Major lupus organ involvement: severe lupus nephritis. Lupus. 2010;19(12):1391–8.
Osio-Salido E, Manapat-Reyes H. Epidemiology of systemic lupus erythematosus in Asia. Lupus. 2010;19(12):1365–73.
Han S, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM, Thomas K, Kaufman KM, Ojwang J, Rojas-Villarraga A, et al. Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE). Hum Mol Genet. 2009;18(6):1171–80.
Deng H, Lu Q, Xu H, Deng X, Yuan L, Yang Z, Guo Y, Lin Q, Xiao J, Guan L, et al. Identification of a novel missense FBN2 mutation in a Chinese family with congenital contractural arachnodactyly using exome sequencing. PLoS One. 2016;11(5):e0155908.
Sengle G, Carlberg V, Tufa SF, Charbonneau NL, Smaldone S, Carlson EJ, Ramirez F, Keene DR, Sakai LY. Abnormal activation of BMP signaling causes myopathy in Fbn2 null mice. PLoS Genet. 2015;11(6):e1005340.
Zhou S, Wang F, Dou Y, Zhou J, Hao G, Xu C, Wang QK, Wang H, Wang P. A novel FBN2 mutation cosegregates with congenital contractural arachnodactyly in a five-generation Chinese family. Clin Case Rep. 2018;6(8):1612–7.
van Loon K, Yemelyanenko-Lyalenko J, Margadant C, Griffioen AW, Huijbers EJM. Role of fibrillin-2 in the control of TGF-beta activation in tumor angiogenesis and connective tissue disorders. Biochim Biophys Acta Rev Cancer. 2020;1873(2):188354.
Yuan Y, Yang M, Wang K, Sun J, Song L, Diao X, Jiang Z, Cheng G, Wang X. Excessive activation of the TLR9/TGF-beta1/PDGF-B pathway in the peripheral blood of patients with systemic lupus erythematosus. Arthritis Res Ther. 2017;19(1):70.
Gardet A, Chou WC, Reynolds TL, Velez DB, Fu K, Czerkowicz JM, Bajko J, Ranger AM, Allaire N, Kerns HM, et al. Pristane-accelerated autoimmune disease in (SWR X NZB) F1 mice leads to prominent tubulointerstitial inflammation and human lupus nephritis-like fibrosis. PLoS One. 2016;11(10):e0164423.
Che R, Motsinger-Reif AA. Evaluation of genetic risk score models in the presence of interaction and linkage disequilibrium. Front Genet. 2013;4:138.
Reid S, Alexsson A, Frodlund M, Morris D, Sandling JK, Bolin K, Svenungsson E, Jonsen A, Bengtsson C, Gunnarsson I, et al. High genetic risk score is associated with early disease onset, damage accrual and decreased survival in systemic lupus erythematosus. Ann Rheum Dis. 2020;79(3):363–9.
Acknowledgements
The technical support group from the Department of Medical Sciences is gratefully acknowledged. We would like to thank the valuable suggestion from Associate Professor Wanling Yang from the Department of Paediatrics and Adolescent Medicine, Queen Mary Hospital, Li Ka Shing Faculty of Medicine, and Centre for Genomic Sciences, Li Ka Shing Faculty of Medicine, and The University of Hong Kong.
Funding
This study was supported by the International Network for Lupus Research (grant no. IRN59W0004), Faculty of Allied Health Sciences, Chulalongkorn University, Fiscal Year 2020 (AHS-CU 63002), Grants for Development of New Faculty Staff, Ratchadaphiseksomphot Endowment Fund, Chulalongkorn University (DNS 63_015_37_002_1), the Faculty of Medicine, Ramathibodi Hospital (CF_61004), and Genomic Thailand funding, Department of Medical Science, Ministry of Public Health. This work was kindly supported by the National Natural Science Foundation of China (Grant No. 81801636).
Author information
Authors and Affiliations
Contributions
NH, PP, and SM conceived the study. WYF and WW performed the genetic data imputation. PT, CT, and PK performed the statistical analysis and function annotation with all figures. NH, PP, JW, SM, DC, and PR provided Thai SLE samples. TS, TC, SK, and NS carried out DNA extraction and genotyping. PT, CT, WYF, and NH wrote the first draft of the manuscript. All authors contributed to the revision of the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All the procedures were followed in compliance with the principles of the Declaration of Helsinki, and informed consent was obtained from all participants. The study and the consent procedures were reviewed and approved by the local institutional review board including the ethical committee from the Faculty of Medicine, Chulalongkorn University (COA no.923/2017), and the Faculty of Medicine, Mahidol University (MURA no. 2015/731, EC no. 590223, Protocol-ID 12-58-12).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Supplementary figure1: The graphs show quantile-quantile plot (Q-Q plot) with inflation factor value in observatory dataset (upper), and replication dataset (lower).
Additional file 2.
Supplementary Table 1: List of significant variants at individual locus from meta-analysis (p-value < 5E-5)
Additional file 3.
Supplementary Table 2: List of HWE of significant variants (p-value < 5E-5)
Additional file 4.
Supplementary figure 2: The regional plots show variants found on ATP6V1B1 (A), ADCY5 (B), DGKG (C), MIR4472-2 (D), and MYO5C allele (E). These SNPs were associated with Thai SLE patients with statistical significant value less than 10E-5.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tangtanatakul, P., Thumarat, C., Satproedprai, N. et al. Meta-analysis of genome-wide association study identifies FBN2 as a novel locus associated with systemic lupus erythematosus in Thai population. Arthritis Res Ther 22, 185 (2020). https://doi.org/10.1186/s13075-020-02276-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13075-020-02276-y