Abstract
Background
Bartonella spp. are vector-borne pathogens transmitted to humans via blood-sucking arthropods. Rodents such as the black rat (Rattus rattus) and Norway rat (R. norvegicus) are thought to be the main reservoirs. An infection with rodent-associated Bartonella spp. may cause severe symptoms in humans such as endocarditis and neuroretinitis. The current knowledge of Bartonella prevalence in rats from western Europe is scarce.
Methods
Rats and a few other rodent by-catches were trapped in the context of a rodenticide resistance study at different sites in Flanders, Belgium. During dissection, biometric data were collected, and spleen tissues were taken. DNA was extracted from spleen samples and tested for Bartonella spp. by conventional generic polymerase chain reaction (PCR). To determine the Bartonella species, a selected number of amplicons were sequenced and compared with GenBank entries.
Results
In total, 1123 rodents were trapped. The predominate species was R. norvegicus (99.64%). Other rodents trapped included: two water voles (Arvicola amphibius, 0.18%); one colour rat (R. norvegicus forma domestica, 0.09%); and one muskrat (Ondatra zibethicus, 0.09%). PCR analysis of 1097 rodents resulted in 410 (37.37%, 95% CI: 34.50–40.31%) Bartonella spp. DNA-positive samples. Bartonella tribocorum (94.68%, 95% CI: 88.02–98.25%) was the most frequently detected Bartonella species, followed by B. grahamii (3.19%, 95% CI: 0.66–9.04%) and B. doshiae (1.06%, 95% CI: 0.03–5.79%). An uncultured Bartonella species occurred in one water vole (1.06%, 95% CI: 0.03–5.79%). There was a significantly higher Bartonella prevalence in older rats compared to juveniles and a significant difference in Bartonella prevalence concerning the localisation of trapping sites. In contrast, there was no statistically significant difference in Bartonella prevalence regarding sex, degree of urbanisation and season.
Conclusions
Based on the high prevalence found, we conclude that the Norway rat seems to be a key reservoir host for zoonotic B. tribocorum in Belgium.

Similar content being viewed by others
Background
The genus Bartonella consists of haemotropic, facultative intracellular, gram-negative α-proteobacteria, which parasitize endothelial cells and erythrocytes of mammalian hosts [1, 2]. Bartonella spp. may cause different diseases such as cat scratch disease, trench fever and Oroya fever with various symptoms, e.g. endocarditis, regional swelling of lymph nodes and vasoproliferative lesions of the skin and abdominal organs [3, 4]. Thus, an undetected infection with these neglected pathogens and inadequate therapy may be life-threatening [5]. Arthropods often play an important role in the transmission of these pathogens; commonly the infectious agents are transmitted to humans via fleas (e.g. the cat flea, Ctenocephalides felis for Bartonella henselae and the rodent flea, Ctenophthalmus nobilis for B. grahamii), the body louse (Pediculus humanus corporis), sand fly (Lutzomyia verrucarum) or ticks (e.g. Ixodes ricinus) [6,7,8,9,10].
Although many different Bartonella species are known, only a few of them are pathogenic to humans [1, 5, 8, 11,12,13,14,15]. Rodents are thought to be the main reservoir for most Bartonella species; however, a majority of these are not zoonotic. The Norway rat (Rattus norvegicus) is known to harbour both non-pathogenic (e.g. B. rattimasiliensis and B. taylorii) as well as pathogenic Bartonella spp. [8, 12, 13]. While rat-associated B. elizabethae, B. vinsonii arupensis and B. washoensis may cause cardiac diseases, B. grahamii is suspected to induce neuroretinitis, and B. tribocorum may cause unspecific symptoms such as fever, apathy and chronic fatigue [8, 12, 13]. Being a reservoir for Bartonella spp. and other zoonotic agents, rodents are crucial for the transmission and maintenance of vector-borne pathogens [7, 9, 16]. In particular, Norway and black rats (R. rattus) may be of concern. As highly synanthropic rodents they inhabit buildings and households, live in close contact to humans in urban and suburban regions and feed on human spoilage [17]. Therefore, monitoring of vector-borne pathogens in connection with rat populations, serving as hosts and reservoirs, is an essential component for the surveillance, prevention and risk control in the context of public health management and One Health politics, especially concerning neglected pathogens such as Bartonella spp. [9, 18].
Many studies worldwide describe moderate to high Bartonella-infection rates in rodents [19, 20]. Seemingly, western Europe is an endemic region with high prevalence levels for Bartonella in rodents and their ectoparasites (France: 11–70% [21,22,23]; Denmark: 30–53% [24]). These studies mostly refer to wild mice and voles. So far, there are only two studies describing Bartonella prevalence rates in rats from western Europe (Marseille, France: 30.3% [21]; Paris, France: 53.5% [25]), whilst another study evaluated the presence of Bartonella spp. in Norway and black rats from different European countries [26]. The latter study also included a pilot investigation of 60 Norway rats from Belgium indicating a high prevalence of B. tribocorum. To further explore the topic of this pilot investigation, the present study aims to: (i) determine the prevalence of Bartonella spp. in Norway rats from Flanders, Belgium; (ii) identify Bartonella species in these rats; and (iii) analyse Bartonella prevalence rates regarding the sex and age of the rats, seasonal influence, geographical location and degree of urbanisation.
Methods
Study sites
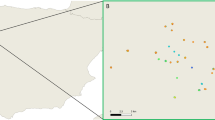
The trapping sites are located in Flanders, the northern part of Belgium (Fig. 1). This region covers an area of 13,682 km2 and is characterized by a mean human population density of 485 individuals/km2 [27,28,29]. Belgium shows classical features of a western industrialised region with well-developed infrastructure as well as agriculture and extensive industry [30]; 97.9% of the population live in urban areas [31]. Only 13.4% of Flanders and Belgium’s capital Brussels are planted with forest, mostly Scots pines (Pinus sylvestris) [27, 32].
Sampling of rodents
In 2015 and 2016 rodents were trapped in the context of a rodenticide resistance study for Norway rats conducted by the Belgian Research Institute for Nature and Forest (INBO, Brussels, Belgium). Detailed information on rodent trapping procedures was published elsewhere [33]. Rodent carcasses were kept frozen (− 20 °C) until necropsies were performed. Biometric data (body length, tail length, body weight and sex) were collected and spleen tissue was taken for further examination. A morphological identification key was used for species identification [34]; for a few individuals, molecular species identification and sex determination was performed. The age classification of rats was based on the body weight: all individuals with a body weight < 200 g were defined as juveniles and all individuals with a body weight ≥ 200 g were classified as adults [35].
DNA extraction
Spleen samples with a size of 1 × 0.5 × 0.2–0.4 cm were homogenized together with 0.6 g sterile ceramic beads (1.4 mm in size; PeqLab Biotechnologie GmbH, Erlangen, Germany) and 500 µl phosphate-buffered saline (PBS, pH 7.2). Homogenization of the tissues was carried out using a Precellys 24 Tissue Homogenizer (PeqLab Biotechnologie GmbH) at 5500 × rpm, twice for 15 s and a break of 10 s in between both runs. Lysis buffer and proteinase K (140 µl and 20 µl, respectively; QIAamp DNA mini kit (Qiagen, Hilden, Germany)) were added to each sample, followed by an overnight incubation at 56 °C in a thermomixer (Eppendorf, Hamburg, Germany). Subsequently, DNA extraction was performed manually with the QIAamp DNA Mini Kit (Qiagen) as recommended by the manufacturer. The quality and quantity of the extracted DNA samples were analysed spectrophotometrically using a NanoDrop ND-1000 (PeqLab Biotechnologie GmbH). Samples with a concentration of > 80 ng/µl DNA were diluted with water (bioscience grade, nuclease-free) to obtain a DNA concentration of 40–80 ng/µl for each sample.
Bartonella spp. DNA detection and rat species identification by conventional PCR and sequence analyses
DNA samples were tested for the presence of Bartonella spp. via conventional polymerase chain reaction (PCR) targeting the gltA gene [36]. Positive samples were further processed by an additional PCR targeting 453–780 bp of the 16S–23S rRNA intergenic spacer (ITS) region [2, 37]. Subsequent electrophoresis was performed for both gene targets on 2% agarose gels stained with HDGreen® Safe DNA Dye (Intas Science Imaging, Göttingen, Germany) and analysed under UV light using a Gel Doc 2000 transilluminator (Bio-Rad Laboratories GmbH, Life Science Group, München, Germany).
A selected number of amplicons of the expected size were further processed for Bartonella species identification. Based on an overview of species determination of randomly chosen Bartonella-positive samples, a selection algorithm was defined. Nineteen pairs of two females (the lightest and the heaviest) and two males (the lightest and the heaviest) were chosen in order to create an overview of sex as well as weight and accordingly age status. Additionally, Bartonella sequence analysis was performed for all positive muskrats (Ondatra zibethicus) and water voles (Arvicolaamphibius).
Selected amplicons were purified manually with the NucleoSpin® Gel and PCR clean-up kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) according to the manufacturer’s instructions, analysed for quality and quantity as mentioned above and commercially sequenced with forward (Ba325s: 5′-CTT CAG ATG ATG ATC CCA AGC CTT CTG GCG-3′) and reverse primers (Ba1100as: 5′-GAA CCG ACG ACC CCC TGC TTG CAA AGC-3′) (Interdisziplinäres Zentrum für Klinische Forschung, Leipzig, Germany). The obtained sequences were aligned and analyzed with BioNumerics (version 7.6; Applied Maths N.V., Sint Martens-Latem, Belgium) and compared with sequences in GenBank using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi). A selection of representative sequences (n = 94) were deposited in the GenBank database under the following accession numbers: MN244575-MN244667.
Molecular species identification was completed for rat samples which could not be identified morphologically. The conventional cytochrome b gene PCR protocols followed the protocols by Parson et al. [38] and Schlegel et al. [39]. Molecular sex determination based on a PCR approach previously described was carried out for a few individuals which were in insufficient condition for morphological sex determination [40, 41]. Visualisation of PCR products and examination of amplicons were performed as indicated above.
GIS analysis: the degree of urbanisation
Geocoordinates (World Geodetic System 1984) were taken during the capture of almost all rats. Hence, geographical information system (GIS) analyses tools were used to analyse the origin of rats (QGIS 3.2.1 ‘Bonn’, Open Source Geospatial Foundation 2019). Census data of human population density (resolution 1 km2) [42] were added and both layers and related information were joined by location. The degree of urbanisation was defined by a population threshold, applied to the population grid cells, as belonging to one of three classes: “urban” (with > 1500 inhabitants/km2); “town” (with 300–1500 inhabitants/km2); and “rural” (with < 300 inhabitants/km2) [43, 44].
Statistical analysis
Confidence intervals (95% CI) with a standard error α = 0.05 for the prevalence of Bartonella spp., sex and species analysis were determined by the Clopper and Pearson method using GraphPad Software (GraphPad Software Inc., SanDiego, CA, USA). Chi-square test (sample size > 30) was conducted for testing the independence of Bartonella prevalence rates concerning sex, season, location and degree of urbanisation. P-values (probability) < 0.05 were considered to be significant and the degrees of freedom (df) were specified by default. Three aspects were analysed in more detail. Individual infection risk as a function of age was assessed using a generalized additive model (gam, binomial error distribution; package gamm4) with a weight smoother function. Additionally, a generalized linear model (glm, lme4 package) with a binomial error distribution was used to evaluate if provinces differed within each class of urbanisation (“urban” was excluded due to the low sample size in each province). Estimated marginal means were calculated using the emmeans package and back-transformed for logit-scale to visualize infection probabilities. Analyses were performed using R software [45]. In addition, geographical foci in the distribution of B. tribocorum genotypes were investigated using a Chi-square test on proportions, where the number of positive samples per state for each genotype would be significantly different to equipartition.
Results
Animal collection
The vast majority of the 1123 trapped rodents were Norway rats (1119/1123; 99.64%, 95% CI: 99.09–99.90%). By-catches represented two water voles (0.18%, 95% CI: 0.02–0.64%), one color rat (Rattus norvegicus f. domestica) (0.09%, 95% CI: < 0.01–0.50%) and one muskrat (0.09%, 95% CI: < 0.01–0.50%).
PCR analyses for Bartonella spp. and analyses of Bartonella prevalence rates in connection with sex, age and degree of urbanisation
A total of 1097 out of 1123 (97.68%, 95% CI: 96.63–98.48%) rodents were tested using the Bartonella PCR. The remaining rats could not be investigated due to the absence of spleen tissue, high grade rotting and/or autolysis. From all 1097 examined rats, 410 (37.37%, 95% CI: 34.50–40.31%) were Bartonella DNA-positive (Table 1). Bartonella DNA-positive samples were distributed all over Flanders (Fig. 1, Additional file 1: Table S1).
As illustrated in Table 1, analysis of the Bartonella prevalence between males and females revealed no statistically significant difference (χ2 = 0.084, df = 1, P = 0.7722).
There was also no significant difference in the prevalence of Bartonella spp. between individuals from rural areas, towns and urban areas (χ2 = 3.167, df = 2, P = 0.2637) and no significant difference was found between seasons regarding Bartonella prevalence rates (χ2 = 3.668, df = 2, P = 0.2254).
However, the prevalence of Bartonella in juvenile rats with a weight < 200 g was significantly lower than in adult rats (χ2 = 11.365, df = 1, P = 0.0007). This is mirrored in the results of gam (Fig. 2), where individual infection risk increases with weight, but remains static at around 260 g before slightly dropping again with increasing weight.
Interestingly, there were statistically significant differences in the prevalence of Bartonella between locations (χ2 = 34.27, df = 4, P < 0.0001). Analysis of differences between provinces for different degrees of urbanisation revealed that this difference was only detected within rural areas. Here, infection probabilities in Antwerp were significantly lower compared to Flemish Brabant, East and West Flanders (Fig. 3). This could not be detected in areas with a higher human population density (town).
Results of the binomial generalized linear model demonstrating probabilities of infection according to location and population density. Focussing on rural areas with a population density < 300 inhabitants/km2, the probability of infection was significantly lower in Antwerp compared with East and West Flanders. This effect could not be demonstrated in areas with a higher human population density such as towns with 300–1500 inhabitants/km2
Sequence analysis of Bartonella-positive samples
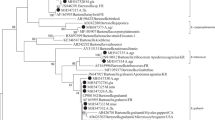
Sequencing of 94 Bartonella-positive samples (94/410; 22.93%, 95% CI: 18.94–27.31%) resulted in the detection of three Bartonella species (Table 2). Bartonella tribocorum (89/94; 94.68%, 95% CI: 88.02–98.25%) was the predominating species. Eighty-eight rodents positive for B. tribocorum were Norway rats and one additional positive animal was a colour rat (Table 2). Bartonella grahamii (3/94; 3.19%, 95% CI: 0.66–9.04%) was detected in one muskrat and two Norway rats, while B. doshiae and uncultured Bartonella sp. were each detected in one water vole (1/94; 1.06%, 95% CI: 0.03–5.79%) (Table 2).
As illustrated in Fig. 4, Norway rats infected with B. tribocorum were found to be broadly distributed. The two B. grahamii DNA-positive Norway rats originated from different parts of Flanders.
Three different B. tribocorum genotypes were defined according to the degree of similarity of 100% (group I), 99% (group II) or 96% (group III) to a sequence (GenBank: HG969192) used as a prototype (Table 3). Bartonella tribocorum genotype I (100% identity to GenBank: HG969192) was normally distributed (χ2 = 2.75, df = 4, P = 0.6005) in the five areas of Flanders (Limburg, Flemish Brabant, Antwerp, East Flanders and West Flanders), whereas genotype II (χ2 = 17.026, df = 4, P = 0.001911) and III (χ2 = 10.444, df = 4, P = 0.03357) exhibited significant differences, cumulating in East and West Flanders (Fig. 5).
Discussion
This study analysed the prevalence of Bartonella spp. in Norway rats from Flanders, Belgium. All rats and a few by-catches were caught in the context of a rodenticide resistance study, which defines rats as a target species in western Europe [33]. The Norway rat is omnipresent in Europe, and since the 20th century has nearly completely displaced the black rat [34].
The overall Bartonella prevalence in the rodent population in this study was 37.4%. This comparatively high prevalence for Bartonella spp. corresponds well with recent data of a pilot study on 60 Norway rats from Belgium with a similarly high Bartonella prevalence (ca. 35% [26]). A lower but also moderate to high prevalence of Bartonella, was described in Norway rats from Europe (France: 30.3% [21]) and other continents, such as North America (Canada: 25% [46]; USA: 25% [47]), South America (Brazil: 19% [48]) and Asia (Taiwan, China: 10.3%, [49]).
Bartonella tribocorum was the most frequently detected species in this study, with 94.7% of all positive samples sequenced. This result is in line with previous findings (89.6% [26]) and suggests that Norway rats are the main reservoir host for B. tribocorum [50], at least in Flanders, Belgium.
Bartonella tribocorum is known to be adapted to rats [8, 12] and to persist in infected erythrocytes without affecting the erythrocytes’ natural life span of about 54–65 days [51]. After the erythrocytes’ apoptosis, B. tribocorum is released into the bloodstream again in order to invade new erythrocytes for replication. Thus, B. tribocorum is able to infect about 1% of the erythrocytes in rats, persisting in the host for a long time without seriously harming the host [52, 53].
There are only a few case reports of B. tribocorum infections in humans, and no reports of bartonellosis in Belgium. Displaying non-specific symptoms such as fever and apathy, Bartonella-infected humans are probably accidental hosts [9, 54]. Until now, there are just a few case reports describing bartonellosis as a zoonosis in central and western Europe. One study refers to six patients in France suffering from different unspecific symptoms; B. tribocorum was detected in two of these patients [55].
In the present study, B. grahamii and B. doshiae occurred in a very small number of samples. However, both species have been frequently reported in Europe [56]. Bartonella grahamii was detected in two R. norvegicus and one O. zibethicus in the present study. These results are in line with findings of B. grahamii in voles and mice from Europe [8, 10, 11]. The occurrence of B. grahamii in rats like R. norvegicus is uncommon but not completely unexpected as this pathogen was previously detected in Norway rats from Taiwan [57]. However, to our knowledge, B. grahamii has not yet been identified in rats from Europe. The occurrence of B. grahamii in O. zibethicus was an unexpected finding, as blood parasites have been thus far rarely found in muskrats [58,59,60]. Further, muskrats are known to be rather insignificant hosts for Bartonella vectors such as ticks, fleas or lice (Pediculus humanus corporis) [61]. They usually act as hosts for hematophagous mites such as Laelaps multispinosa, Zibethacarus ondatrae and Listrophorus spp. [62, 63]. Possible reasons for the very low infestation with ectoparasites are the predominantly aquatic habitat of muskrats and their very dense fur [60]. To the best of our knowledge, this is the first detection of Bartonella species in a muskrat. As noted above, all Bartonella spp. are rather host-specific, with some Bartonella spp. being more host-specific than others. Host specificity of Bartonella spp. is closely related associated with the ability of adhering to and invading a host cell [64].
Bartonella grahamii possesses a higher number of genes for host-adaptability than human-associated bartonellae. These genes are located in a dynamic region being involved in a phage-derived run-off replication. This results in a high number of amplified genes associated with host-adaptation and a rapid diversification of these genes, enabling rodent-associated Bartonella species like B. grahamii to host shifts [64, 65]. Perhaps that is why B. grahamii was detected in a muskrat and is adapted to various host species such as Microtus spp., Myodes spp. or Apodemus spp. [8]. Furthermore, B. grahamii is a zoonotic agent causing more specific and severe symptoms such as neuroretinitis [8, 13].
Until now, B. doshiae has not been associated with clinical human cases and thus is considered as non-pathogenic to humans [8]. Bartonella doshiae is a common European Bartonella species known to infect different Rattus species [66], voles of the genus Microtus [8, 56] and bank voles (Myodes glareolus [67, 68]). Hence, the occurrence of B. doshiae in one water vole was not unexpected, although prevalence studies of Bartonella spp. in water voles are lacking.
Three different sequence types I, II and III with 96%, 99%, 100% identity, respectively to the 16S-23S rRNA ITS region reference sequence of B. tribocorum (GenBank: HG969192) were detected. Individuals with sequence types II and III cumulated in the western provinces of Flanders (East Flanders and West Flanders; see Fig. 3). Whereas the knowledge about different sequence types in B. tribocorum is scarce, far more is known about sequence types (ST) in B. henselae. There are 118 different B. henselae strains, which were classified into 12 to 14 STs [69, 70]. Arvand et al. [70] confirmed the association between different STs of B. henselae and different hosts: most human pathogenic B. henselae isolates (66%) from Europe were identified as ST1, while most feline isolates (27.2%) belong to ST7. Sequence types, however, differed regionally between the UK and Spain (UK, ST2; Spain, ST1) in humans [69, 71].
The occurrence and host specificity of the sequence types of B. henselae seem to depend on the geographical location. Besides, coevolutionary processes have been observed between Bartonella spp. and rodent hosts [72]. Possibly the accumulation of the sequence types II and III in West Flanders results from geographical distribution patterns or from a coevolution with the mammalian host. A combination of the two seems also possible.
Moreover, we found that the sex of rodents had no influence on the prevalence of Bartonella. Kosoy et al. [73] came to the same conclusion, examining the relationship of Bartonella prevalence and sex in a wild cotton rat (Sigmodon hispidus) population from Georgia, USA. In contrast, the age of rats plays an important role for the individual infection status with Bartonella in Norway rats. In the present study, juvenile animals were significantly less frequently Bartonella DNA-positive (Table 1 and Fig. 4). In contrast to our result, investigations of wild cotton rats and American bush rats (Neotoma micropus and N. albigula) showed that juveniles had a significantly higher prevalence of Bartonella than adults [73, 74]. Additionally, a study of small mammals (Myodes spp., Apodemus spp., Microtus spp. and Sorex spp.) in central Europe concluded that there is no significant effect of age on individual infection probability in any of the small mammal species [20]. However, those studies investigated different species and the methodology differed from the present study.
A possible explanation as to why older animals are significantly more likely to be Bartonella DNA-positive could be found in the infectious life-cycle of B. tribocorum. This species can persist in the host until its death without fatally damaging it [52, 53].
In our study, no significant differences in Bartonella prevalence rates according to season were found, although comparable studies demonstrate those for cotton rats from the USA where a very high infection level was reached in autumn (95%) with a lower prevalence in early summer (49%) [73]. This discrepancy might be explained by the distinct families of tested rodents, different study design and in particular by the absence of rat trapping in the autumn in our study.
Statistical analysis revealed the province of Antwerp exhibiting a significantly lower prevalence of Bartonella spp. in rural areas compared to East and West Flanders. The reason behind this lower prevalence remains speculative. However, the province of Antwerp, with a gross domestic product of € 80,981 million (2016, Data Explorer [75]), is one of Flanders most powerful economic and industrial regions with consequences on natural habitats and the absence of vectors, particularly in rural areas.
The retrospective data analysis of Bartonella prevalence according to degree of urbanisation was limited (unbalanced group sizes). Nevertheless, the results of this study are in line with Obiegala et al. [26], who demonstrated no significant differences of Bartonella prevalence according to the human population density.
Although other authors have described a high prevalence of Bartonella spp. (53.5%) in an urban Norway rat population from western Europe (France, Paris [25]), we detected a lower, but still quite high, prevalence of Bartonella spp. (37.4%).
The group size of urban rats was the smallest compared to the town and rural rats but is of particular importance for the transmission of zoonoses [17]. Hence, urban rats live in close vicinity to humans and act as reservoirs for many other human pathogenic agents such as Yersinia enterocolitica, Y. pseudotuberculosis and Y. pestis, as well as Leptospira interrogans, Rickettsia typhi, Streptobacillus moniliformis and Seoul orthohantavirus [33, 76].
Conclusions
An infection with Bartonella spp. is rarely diagnosed in European countries and bartonellosis became one of Europe’s most neglected diseases. However, the results of the present study and other studies [20,21,22,23,24] reinforce that Bartonella spp. pose a real threat to human health. The high overall prevalence of Bartonella spp. (37.4%) reported here proves that there are human pathogenic Bartonella species in rats from Belgium. Our results suggest that rats of the species R. norvegicus seem to be a major reservoir host for Bartonella spp., especially for B. tribocorum, in Flanders, Belgium.
Availability of data and materials
The data supporting the findings of this study are included within this article and its additional file. Raw datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. Representative sequences were submitted to the GenBank database under the accession numbers MN244575-MN244667.
Abbreviations
- BLASTn:
-
Basic Local Alignment Search Tool for nucleotides
- bp:
-
base pairs
- CI:
-
confidence interval
- df:
-
degrees of freedom
- DNA:
-
deoxyribonucleic acid
- gam:
-
generalized additive model
- GIS:
-
geographical information system
- ITS:
-
intergenic spacer
- PBS:
-
phosphate-buffered saline
- PCR:
-
polymerase chain reaction
- rpm:
-
rounds per minute
- rRNA:
-
ribosomal ribonucleic acid
- ST:
-
Sequence type
- UV:
-
Ultraviolet
References
Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet Res. 2005;36:383–410.
Schorn S, Pfister K, Reulen H, Mahling M, Silaghi C. Occurrence of Babesia spp., Rickettsia spp. and Bartonella spp. in Ixodes ricinus in Bavarian public parks, Germany. Parasit Vectors. 2011;4:135.
Relman DA, Loutit JS, Schmidt TM, Falkow S, Tompkins LS. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–80.
Koehler JE, Tappero JW. Bacillary angiomatosis and bacillary peliosis in patients infected with human immunodeficiency virus. Clin Infect Dis. 1993;17:612–24.
Prutsky G, Domecq JP, Mori L, Bebko S, Matzumura M, Sabouni A, et al. Treatment outcomes of human bartonellosis: a systematic review and meta-analysis. Int J Infect Dis. 2013;17:e811–9.
Brinkerhoff RJ, Kabeya H, Inoue K, Bai Y, Maruyama S. Detection of multiple Bartonella species in digestive and reproductive tissues of fleas collected from sympatric mammals. ISME J. 2010;4:955–8.
Rizzoli A, Silaghi C, Obiegala A, Rudolf I, Hubálek Z, Földvári G, et al. Ixodes ricinus and its transmitted pathogens in urban and peri-urban areas in Europe: new hazards and relevance for public health. Front Public Health. 2014;2:251.
Vayssier-Taussat M, Le Rhun D, Bonnet S, Cotté V. Insights in Bartonella host specificity. Ann N Y Acad Sci. 2009;1166:127–32.
Tomassone L, Berriatua E, de Sousa R, Duscher GG, Mihalca AD, Silaghi C, et al. Neglected vector-borne zoonoses in Europe: into the wild. Vet Parasitol. 2018;251:17–26.
Silaghi C, Pfeffer M, Kiefer D, Kiefer M, Obiegala A. Bartonella, rodents, fleas and ticks: a molecular field study on host-vector-pathogen associations in Saxony, eastern Germany. Microb Ecol. 2016;72:965–74.
Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. Bartonella spp. in pets and effect on human health. Emerg Infect Dis. 2006;12:389–94.
Ellis BA, Regnery RL, Beati L, Bacellar F, Rood M, Glass GG, et al. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–4.
Inoue K, Maruyama S, Kabeya H, Yamada N, Ohashi N, Sato Y, et al. Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl Environ Microbiol. 2008;74:5086–92.
Maggi RG, Mozayeni BR, Pultorak EL, Hegarty BC, Bradley JM, Correa M, Breitschwerdt EB. Bartonella spp. bacteremia and rheumatic symptoms in patients from Lyme disease-endemic region. Emerg Infect Dis. 2012;18:783–91.
Maguiña C, Gotuzzo E. Bartonellosis. Infect Dis Clin N Am. 2000;14:1–22.
Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Infect Dis. 2005;11:1842–7.
Feng AYT, Himsworth CG. The secret life of the city rat: a review of the ecology of urban Norway and black rats (Rattus norvegicus and Rattus rattus). Urban Ecosyst. 2014;17:149–62.
Cunningham AA, Daszak P, Wood JLN. One health, emerging infectious diseases and wildlife: two decades of progress? Philos Trans R Soc Lond B Biol Sci. 2017;19:372.
Gutiérrez R, Krasnov B, Morick D, Gottlieb Y, Khokhlova IS, Harrus S. Bartonella infection in rodents and their flea ectoparasites: an overview. Vector Borne Zoonotic Dis. 2015;15:27–39.
Obiegala A, Jeske K, Augustin M, Król N, Fischer S, Mertens-Scholz K, et al. Highly prevalent bartonellae and other vector-borne pathogens in small mammal species from the Czech Republic and Germany. Parasit Vectors. 2019;12:332.
Gundi VAKB, Davoust B, Khamis A, Boni M, Raoult D, La Scola B. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol. 2004;42:3816–8.
Buffet JP, Pisanu B, Brisse S, Roussel S, Félix B, Halos L, et al. Deciphering Bartonella diversity, recombination, and host specificity in a rodent community. PLoS ONE. 2013;8:e68956.
Buffet JP, Marsot M, Vaumourin E, Gasqui P, Masséglia S, Marcheteau E, et al. Co-infection of Borrelia afzelii and Bartonella spp. in bank voles from a suburban forest. Comp Immunol Microbiol Infect Dis. 2012;35:583–9.
Engbaek K, Lawson PA. Identification of Bartonella species in rodents, shrews and cats in Denmark: detection of two B. henselae variants, one in cats and the other in the long-tailed field mouse. APMIS. 2004;112:336–41.
Desvars-Larrive A, Pascal M, Gasqui P, Cosson JF, Benoît E, Lattard V, et al. Population genetics, community of parasites, and resistance to rodenticides in an urban brown rat (Rattus norvegicus) population. PLoS ONE. 2017;12:e0184015.
Obiegala A, Heuser E, Ryll R, Imholt C, Fürst J, Prautsch LM, et al. Norway and black rats in Europe: potential reservoirs for zoonotic arthropod-borne pathogens. Pest Manag Sci. 2019;75:1556–63.
van Landuyt W, Vanhecke L, Hoste I, Bauwens D. Do the distribution patterns of vascular plant species correspond to biogeographical classifications based on environmental data? A case study from northern Belgium. Landsc Urban Plan. 2011;99:93–103.
Strubbe D, Matthysen E. Predicting the potential distribution of invasive ring-necked parakeets Psittacula krameri in northern Belgium using an ecological niche modelling approach. Biol Invasions. 2009;11:497–513.
Bevolking: omvang en groei. https://www.statistiekvlaanderen.be/bevolking-omvang-en-groei#in_de_vlaamse_ruit_wonen_meeste_inwoners_per_km%C2%B2. Accessed 19 Apr 2019.
Maes D, van Dyck H. Butterfly diversity loss in Flanders (north Belgium): Europeʼs worst case scenario? Biol Conserv. 2001;99:263–76.
Belgium - urbanization 2016. Statistic. https://www.statista.com/statistics/455783/urbanization-in-belgium/. Accessed 9 July 2018.
Verheyen K, Vanhellemont M, Stock T, Hermy M. Predicting patterns of invasion by black cherry (Prunus serotina Ehrh) in Flanders (Belgium) and its impact on the forest understorey community. Divers Distrib. 2007;13:487–97.
Rouffaer LO, Baert K, van den Abeele AM, Cox I, Vanantwerpen G, de Zutter L, et al. Low prevalence of human enteropathogenic Yersinia spp. in brown rats (Rattus norvegicus) in Flanders. PLoS ONE. 2017;12:e0175648.
Westheide W, Rieger G. Spezielle Zoologie. Teil 2: Wirbel- oder Schädeltiere: Spektrum Akademischer Verlag; 2009.
Webster JP, Ellis WA, Macdonald DW. Prevalence of Leptospira spp. in wild brown rats (Rattus norvegicus) on UK farms. Epidemiol Infect. 1995;1995:195–201.
Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–803.
Maggi RG, Breitschwerdt EB. Potential limitations of the 16S-23S rRNA intergenic region for molecular detection of Bartonella species. J Clin Microbiol. 2005;43:1171–6.
Parson W, Pegoraro K, Niederstätter H, Föger M, Steinlechner M. Species identification by means of the cytochrome b gene. Int J Legal Med. 2000;114:23–8.
Schlegel M, Ali HS, Stieger N, Groschup MH, Wolf R, Ulrich RG. Molecular identification of small mammal species using novel cytochrome b gene-derived degenerated primers. Biochem Genet. 2012;50:440–7.
Aasen E, Medrano JF. Amplification of the Zfy and Zfx genes for sex identification in humans, cattle, sheep and goats. Biotechnology. 1990;8:1279.
Bryja J, Konecny A. Fast sex identification in wild mammals using PCR amplification of the Sry gene. Folia Zoologica. 2003;52:269–74.
European Commission EU. GEOSTAT—Eurostat. https://ec.europa.eu/eurostat/web/gisco/geodata/reference-data/population-distribution-demography/geostat#geostat11. Accessed 31 July 2019.
Heuser E, Fischer S, Ryll R, Mayer-Scholl A, Hoffmann D, Spahr C, et al. Survey for zoonotic pathogens in Norway rat populations from Europe. Pest Manag Sci. 2017;73:341–8.
Serrano Giné D, Russo A, Brandajs F, Pérez Albert MY. Characterizing European urban settlements from population data: a cartographic approach. Cartogr Geogr Inf Sci. 2016;43:442–53.
R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for statistical computing; 2018.
Himsworth CG, Bai Y, Kosoy MY, Wood H, DiBernardo A, Lindsay R, et al. An investigation of Bartonella spp., Rickettsia typhi, and Seoul hantavirus in rats (Rattus spp.) from an inner-city neighborhood of Vancouver, Canada: is pathogen presence a reflection of global and local rat population structure? Vector Borne Zoonotic Dis. 2015;15:21–6.
Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio. 2014;5:e01933-14.
Costa F, Porter FH, Rodrigues G, Farias H, de Faria MT, Wunder EA, et al. Infections by Leptospira interrogans, Seoul virus, and Bartonella spp. among Norway rats (Rattus norvegicus) from the urban slum environment in Brazil. Vector Borne Zoonotic Dis. 2014;14:33–40.
Lin JW, Chen CY, Chen WC, Chomel BB, Chang CC. Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J Med Microbiol. 2008;57:1496–501.
Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, et al. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–9.
Derelanko MJ. Determination of erythrocyte life span in F-344, Wistar, and Sprague-Dawley rats using a modification of the ([3H]DFP) method. Fundam Appl Toxicol. 1987;9:271–6.
Schülein R, Seubert A, Gille C, Lanz C, Hansmann Y, Piémont Y, Dehio C. Invasion and persistent intracellular colonization of erythrocytes. J Exp Med. 2001;193:1077–86.
Harms A, Dehio C. Intruders below the radar: molecular pathogenesis of Bartonella spp. Clin Microbiol Rev. 2012;25:42–78.
Breitschwerdt EB, Kordick DL. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev. 2000;13:428–38.
Vayssier-Taussat M, Moutailler S, Féménia F, Raymond P, Croce O, La Scola B, et al. Identification of novel zoonotic activity of Bartonella spp., France. Emerg Infect Dis. 2016;22:457–62.
Oliver MK, Telfer S, Piertney SB. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc Biol Sci. 2009;276:1119–28.
Hsieh JW, Tung KC, Chen WC, Lin JW, Chien LJ, Hsu YM, et al. Epidemiology of Bartonella infection in rodents and shrews in Taiwan. Zoonoses Public Health. 2010;57:439–46.
Warwick T. The parasites of the muskrat (Ondatra zibethica L.) in the British Isles. Parasitology. 1936;28:395.
Grundmann AW, Tsai YH. Some parasites of the muskrat, Ondatra zibethicus osoyoosensis (Lord, 1863) Miller, 1912 from the Salt Lake Valley, Utah. Trans Am Microsc Soc. 1967;86:139.
Cross HB, Campbell-Palmer R, Girling S, Rosell F. The Eurasian beaver (Castor fiber) is apparently not a host to blood parasites in Norway. Vet Parasitol. 2012;190:246–8.
Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, Birtles RJ, et al. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet Res. 2009;40:29.
Bauer CA, Whitaker JO. Ectoparasites of muskrats from Indiana with special emphasis on spatial distribution of coexisting mites of the genus Listrophorus. Am Midl Nat. 1981;105:112.
Rockett CL, Johnston SA. Ectoparasitic arthropods collected from some northern Ohio mammals. Great Lakes Entomol. 1988;21:147–9.
Vayssier-Taussat M, Le Rhun D, Deng HK, Biville F, Cescau S, Danchin A, et al. The Trw type IV secretion system of Bartonella mediates host-specific adhesion to erythrocytes. PLoS Pathog. 2010;6:e1000946.
Berglund EC, Frank AC, Calteau A, Vinnere Pettersson O, Granberg F, Eriksson AS, et al. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet. 2009;5:e1000546.
Angelakis E, Raoult D. Pathogenicity and treatment of Bartonella infections. Int J Antimicrob Agents. 2014;44:16–25.
Birtles RJ, Hazel SM, Bennett M, Bown K, Raoult D, Begon M. Longitudinal monitoring of the dynamics of infections due to Bartonella species in UK woodland rodents. Epidemiol Infect. 2001;126:323–9.
Schmidt S, Essbauer SS, Mayer-Scholl A, Poppert S, Schmidt-Chanasit J, Klempa B, et al. Multiple infections of rodents with zoonotic pathogens in Austria. Vector Borne Zoonotic Dis. 2014;14:467–75.
Chaloner GL, Harrison TG, Coyne KP, Aanensen DM, Birtles RJ. Multilocus sequence typing of Bartonella henselae in the United Kingdom indicates that only a few, uncommon sequence types are associated with zoonotic disease. J Clin Microbiol. 2011;49:2132–7.
Arvand M, Feil EJ, Giladi M, Boulouis HJ, Viezens J. Multi-locus sequence typing of Bartonella henselae isolates from three continents reveals hypervirulent and feline-associated clones. PLoS ONE. 2007;2:e1346.
Gil H, Escudero R, Pons I, Rodríguez-Vargas M, García-Esteban C, Rodríguez-Moreno I, et al. Distribution of Bartonella henselae variants in patients, reservoir hosts and vectors in Spain. PLoS ONE. 2013;8:e68248.
Lei BR, Olival KJ. Contrasting patterns in mammal-bacteria coevolution: Bartonella and Leptospira in bats and rodents. PLoS Negl Trop Dis. 2014;8:e2738.
Kosoy M, Mandel E, Green D, Marston E, Childs J. Prospective studies of Bartonella of rodents. Part I. Demographic and temporal patterns in population dynamics. Vector Borne Zoonotic Dis. 2004;4:285–95.
Morway C, Kosoy M, Eisen R, Montenieri J, Sheff K, Reynolds PJ, Powers N. A longitudinal study of Bartonella infection in populations of woodrats and their fleas. J Vector Ecol. 2008;33:353–64.
Eurostat—Data Explorer. http://appsso.eurostat.ec.europa.eu/nui/show.do?dataset=nama_10r_3gdp&lang=de. Accessed 27 Oct 2018.
Himsworth CG, Jardine CM, Parsons KL, Feng AYT, Patrick DM. The characteristics of wild rat (Rattus spp.) populations from an inner-city neighborhood with a focus on factors critical to the understanding of rat-associated zoonoses. PLoS ONE. 2014;9:e91654.
Acknowledgements
Special thanks to Dana Rüster, Nadja Leinecker, Evelin Brumme and Mario Reinhardt for excellent technical assistance, to Dörte Kaufmann and Elisa Heuser for molecular species and sex determination of some Norway rats, and to Stephanie Speck, Johanna Fürst, Lisa Eisenlöffel and Yauhen Karliuk for organisational support.
Funding
The funding was provided by institutional funds of the Institute of Animal Hygiene and Veterinary Public Health of the University of Leipzig. RGU acknowledges support by DZIF (TTU "Emerging Infections").
Author information
Authors and Affiliations
Contributions
AO, MP and MK elaborated the study concept and design. KB, MK and AO collected the data. MK, AO and NK performed the experiments. CI and MK performed data analysis and visualization. MK wrote a draft of the manuscript. AO, MP, NK, RGU, CI and KB reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Rat trapping was performed by trained pest controllers with special technical knowledge. Norway rats are regarded as a major pest species in Europe. In order to control the population in Flanders, Belgium, trapping and killing of the rats did not require a legal permission. Other captured species mentioned below were presumed to be by-catch. Detailed information has been published elsewhere [33].
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
PCR results for Bartonella spp. and trapping number with coordinates of trapping sites (N, North; E, East) per collected rodent, Belgium, 2015–2016.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Krügel, M., Pfeffer, M., Król, N. et al. Rats as potential reservoirs for neglected zoonotic Bartonella species in Flanders, Belgium. Parasites Vectors 13, 235 (2020). https://doi.org/10.1186/s13071-020-04098-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04098-y