Abstract
Background
Fasciola gigantica, the tropical liver fluke, infects buffaloes in Asian and African countries and causes significant economic losses and poses public health threat in these countries. However, little is known of the transcriptional response of buffaloes to infection with F. gigantica. The objective of the present study was to perform the first transcriptomic analysis of buffalo liver infected by F. gigantica. Understanding the mechanisms that underpin F. gigantica infection in buffaloes will contribute to our ability to control this parasite.
Methods
We challenged buffaloes with 500 viable F. gigantica metacercariae and collected liver samples through a time course at 3, 42 and 70 days post-infection (dpi). Then, we performed gene expression analysis on liver samples using RNA sequencing (RNA-Seq) Illumina technology and confirmed the RNA-Seq data by quantitative RT-PCR analysis.
Results
Totals of 496, 880 and 441 differentially expressed transcripts were identified in the infected livers at 3, 42 and 70 dpi, respectively. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that transcriptional changes in the liver of infected buffaloes evolve over the course of infection. The predominant response of buffaloes to infection was mediated by certain pathways, such as MHC antigen processing and presentation, Toll-like receptor 4 (TLR4), transforming growth factor beta (TGF-β), and the cytochrome P450. Hepatic drug metabolizing enzymes and bile secretion were also affected.
Conclusions
Fasciola gigantica can induce statistically significant and biologically plausible differences in the hepatic gene expression of infected buffaloes. These findings provide new insights into the response of buffaloes to F. gigantica over the course of infection, which may be useful in determining pathways that can modulate host-parasite interaction and thus potentially important for clearance of the parasite.
Similar content being viewed by others
Background
Fasciolosis is a serious liver disease caused by infection with the digenetic trematodes Fasciola hepatica and F. gigantica in temperate and tropical countries, respectively [1]. These flukes enter the definitive host, such as cattle, buffaloes, sheep and goats, orally and migrate towards the liver via the peritoneal cavity. Clinically affected animals exhibit a reduction in the growth rates, development and productivity, and in severe cases fasciolosis may lead to death [2]. Fasciolosis causes significant economic losses in livestock industry [3] and is a serious public health problem [4] by causing liver fibrosis, cirrhosis and cancer in humans [5]. Adult flukes have been recovered from the bile duct of humans from almost all continents [6, 7]. Reported estimates indicate that up to 17 million people are infected worldwide and that about 91 million are at risk [8].
Fasciola gigantica (tropical liver fluke), the major fluke infecting ruminants in Asia and Africa, can adversely affect the weight gain, feed conversion efficiency and reproduction of the affected buffaloes, imposing a serious threat to buffalo farming in these countries [9]. Current methods to control liver fluke infection rely on the use of fasciocidal drugs. However, the escalating anthelmintic resistance (AR) in ruminants has become a major concern [10, 11]. In an effort to discover novel alternative fasciocidal drugs, the anthelmintic efficacy of some medicinal plants against liver flukes have been tested [12, 13]. Also, many immunization trials in mice [14], rabbits [15], sheep [16, 17], goats [18] and cattle [19] exploiting various antigens and adjuvant systems [20] have been reported, but unfortunately these trials did not evoke adequate immune response to protect against challenge infection [18].
The liver fluke F. hepatica infection induces a dominant Th2/T-regulatory type immune response [21] and is known to modulate the host immune responses by various mechanisms, including the production of immune-suppressive cytokines and the alternative activation of macrophages [22], the increase of regulatory T cells [23] and the modulation of differentiation and function of dendritic cells [24, 25]. In contrast, the immunity elicited against F. gigantica infection is a mix of Th1/Th2 response with a predominance of a Th2-biased pattern [26, 27]. Recent studies have employed RNA sequencing (RNA-seq) to elucidate the expressions of genes associated with host’s immune responses, metabolism and transcriptomic changes in peripheral blood mononuclear cells (PBMCs) [28–30]. However, much still is unknown regarding the host immune response mechanisms against F. gigantica and the extent to which this response contributes to the resilience of buffaloes compared to certain cattle breeds [31, 32].
Herein, we utilized RNA-Seq to determine the transcriptional profiles of the liver of buffaloes infected with F. gigantica. We compared the differential gene expression of liver from infected buffaloes to that from uninfected controls. As the first report of a transcriptome analysis of buffalo liver during F. gigantica infection, the data presented here provide new insights into the response of buffalo to F. gigantica and revealed distinct pathways that are dysregulated by F. gigantica in the liver of infected buffaloes.
Methods
Metacercariae
Eggs of F. gigantica were collected from the gall-bladder of naturally infected buffaloes from Guangxi Zhuang Autonomous Region, PR China, and incubated at 29 °C for 11 days. The newly-hatched miracidia were used to infect Galba pervia snails (3–5 miracidia/snail) maintained in tissue culture plastic plate for 2 h and then infected snails were incubated in order to allow the miracidium stage to develop to sporocyst, redia, daughter redia and finally to cercariae. After 42 days, fully-developed cercariae emerged from the snails and were harvested and developed into metacercariae on 5 × 5 cm cellophane sheets. The metacercariae on cellophane sheets were washed several times with phosphate buffered saline (PBS) and were used immediately to infect buffaloes as described previously [33].
Animals and experimental infection
Eighteen buffaloes, 8–10-month-old, were purchased from a water buffalo farm in Guangxi Zhuang Autonomous Region, PR China. Animals were randomly divided into two groups: (i) nine for the non-infected control group and (ii) nine for the infected group. Each group was further subdivided into 3 subgroups, each of 3 buffaloes. To rule out any prior infection with F. gigantica, faecal examination and ELISA testing using IgG and IgM antibodies against F. gigantica were performed [34]. Also, after an acclimatization period of 2 weeks, all buffaloes were treated with triclabendazole 5% w/v oral suspension in order to eliminate any liver flukes. After four weeks of the withdrawal time, nine buffaloes were infected orally with 500 viable metacercariae per animal, whereas control animals were mock-inoculated with 0.85% NaCl solution without metacercariae [31]. At 3, 42 and 70 days post-infection (dpi), three animals from each of the infected and control groups were sacrificed and their livers were collected and stored at -80 °C until analysis. Liver was selected because it is the target organ and preferable habitat of F. gigantica flukes in the definitive host (buffaloes) and in the mean time it performs many vital physiological, metabolic and immunological functions in the body [35]. At necropsy, whole blood samples of all animals were collected aseptically into tubes without anticoagulant and were separated by centrifugation for collection and testing of the sera. Fasciola gigantica infection was also confirmed by observing gross pathological lesions associated with the presence of adult flukes in the liver of infected animals.
RNA preparation
Total RNA was extracted from individual liver samples using Trizol reagent according to the manufacturer’s instructions (Invitrogen Co. Ltd, San Diego, USA). All RNA samples were treated with 20 units of RQ1 RNase-Free DNase (Promega, Madison, USA) to remove any residual genomic DNA according to the manufacturer’s protocol. Agilent Bioanalyzer 2100 (Agilent Technologies, CA, USA) and NanoPhotometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA) were used to evaluate the integrity and purity of RNA samples, respectively.
Library preparation, clustering and transcriptome sequencing
RNA (3 μg) of each liver sample was used for the preparation of RNA libraries. Eighteen sequencing libraries were constructed using NEBNext® Ultra™ RNA LibraryPrep Kit for Illumina® (NEB, Ipswich, MA, USA) following the manufacturer’s protocol and index codes were added to attribute the sequences to the corresponding sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In order to select cDNA fragments ranging from 150 bp to 200 bp, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Three μl USER Enzyme (NEB, Ipswich, MA, USA) were used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. Then, the PCR was carried out with Phusion High-Fidelity DNA polymerase, universal PCR primers and index (X) Primer. PCR products were purified (AMPure XP system) and library quality was evaluated on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions.
Data analysis
Raw reads of fastq format were processed through in-house Perl scripts. Clean reads were obtained by removing reads adapters, ploy-N containing reads and low quality reads from raw data. At the same time, Q20, Q30 and GC content of clean data were calculated. All downstream analyses were based on the clean data with high quality. Bubalus bubalis genome was used as the reference genome and gene model annotation files were downloaded from the water buffalo genome website (http://www.ncbi.nlm.nih.gov/genome/?term=Bubalus+bubalis). Index of the reference genome was built using Bowtie v2.2.3 and paired-end clean reads were aligned to Bubalus bubalis reference genome using TopHat v2.0.12. We selected TopHat as the mapping tool because it can generate a database of splice junctions based on the gene model annotation file and provide a better mapping result than other non-splice mapping tools [36]. HTSeq v0.6.1 was used to count the read’s numbers mapped to each gene. Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced (FPKM) of each gene was calculated for estimating gene expression levels. Differential expression analysis of two groups (three replicates per group) was performed using the DESeq R package (1.18.0) [37]. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Gene expression differences were considered significant if the adjusted P-value was < 0.05 and > 1.5-fold change was observed in expression levels.
Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analysis
Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) was implemented using the GOseq R package [38]. GO enrichment analysis was performed by collating all the GO terms that were significantly enriched in the identified DEG, and followed by filtering the DEGs based on the biological functions. All DEGs were mapped to GO terms in the database (http://www.geneontology.org/), and then gene numbers were calculated for every term using the hypergeometric test in order to obtain significantly enriched GO terms for DEGs; these were compared to the genomic background. GO terms with corrected P-value less than 0.05 were considered significantly enriched by DEGs. KOBAS software was used to perform pathway enrichment analysis and to test the statistical enrichment of the DEGs in KEGG (http://www.genome.jp/kegg/) [39, 40]. This analysis was used to identify significant enrichment of genes involved in metabolic or signalling pathways. AnimalTFDB (http://www.bioguo.org/AnimalTFDB/) was employed to identify and classify the transcriptional factors (TFs) in the genome of water buffalo.
qRT-PCR verification of RNA-Seq expression patterns
Total RNA was isolated from infected livers and non-infection controls at 3, 42 and 70 dpi using RNeasy Mini Kit (QIAGEN Gmbh, Hilden, Germany). DNase-digested total RNA (1 μg) was reverse-transcripted to single strand cDNA using the RT2 First Strand Kit (QIAGEN Science, Maryland, USA) according to the manufacturer's protocol. RT2 SYBR® Green ROX qPCR Mastermix (QIAGEN Gmbh, Hilden, Germany) was used to perform qRT-PCR reaction on ABI’s real-time PCR cycler, the ABI 7500, according to the manufacturer’s instructions. Eleven genes were randomly selected for qRT-PCR verification. Forward (F) and reverse (R) primer pairs used to amplify genes of interest in the qRT-PCR reactions are listed in Table 1. The amplification reactions were performed using following conditions: 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 60 °C for 1 min. Melting curve analysis was carried out using following conditions: 1 min at 95 °C, 65 °C for 2 min and progressive increase from 65 °C to 95 °C to ensure that a single product was amplified in each reaction.
Results
Confirmation of F. gigantica infection in buffaloes
Fasciola gigantica infection was confirmed in all challenged buffaloes by observing gross pathological lesions and adults F. gigantica flukes (Additional files 1 and 2: Figure S1 and Figure S2). Livers of the control uninfected buffaloes appeared normal and were free of any F. gigantica flukes. Serological testing using ELISA confirmed the infection in all animals challenged with metacercariae at 42 and 70 dpi.
Transcriptomic features of buffalo livers following F. gigantica infection
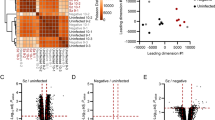
Over 58,000,000 raw reads (Additional file 3: Table S1) were obtained from each liver sample. The RNA-seq raw data are available at NCBI (accession no: PRJNA341921). More than 53,000,000 clean reads were obtained after removing low quality reads and adaptors. More than 70% clean reads were distributed in exon regions and the rest in introns or intergenics. A total of 496, 880 and 441 transcripts were identified as differentially expressed at 3, 42 and 70 dpi, respectively, compared to non-infected control groups (Fig. 1). The RNA-seq results were confirmed by qRT-PCR (Fig. 2). This analysis revealed five DEGs in all animal groups including butyrophilin subfamily 1 member A1-like (Gene ID: 102404197), myeloid-associated differentiation marker-like (Gene ID: 102406172), phosphoserine aminotransferase 1 (Gene ID: 102410803), and two new genes (Fig. 3).
Volcano map of the differentially expressed genes between infected and control buffaloes. Significantly differentially expressed genes are shown as red (up) or green (down) dots. No significant difference between the expressions of genes is indicated by blue dots. Ordinate represents the magnitude of gene expression changes. The x-axis represents the value of log2(fold change) and the y-axis shows the value of -log10(pval). a, b and c represent differentially expressed genes at 3, 42 and 70 days post-infection, respectively
GO classification
GO enrichment analysis (www.geneontology.org) revealed top 30 significant differentially expressed GO terms that were classified into “molecular function”, “biological process”, and “cellular component” as shown in Fig. 4. At 3 dpi, several GO terms classified in biological process showed upregulation. Only two significant GO terms were classified under “cellular component”, including “extra chromosomal circular DNA” and “extra chromosomal DNA”. While at 42 dpi, top 30 significant differentially expressed GO terms were only classified into “molecular function” and “biological process”. The “immune response”, “immune system process” and “cytokine activity” were significantly enriched at 70 dpi.
Differentially expressed GO terms. Differentially expressed genes (DEGs) were classified into three main categories: molecular function, cellular component and biological process. The identified functions and the corresponding numbers of DEGs for each GO category are shown. a Top 30 DE molecular function, cellular component and biological process in A2T (infected) vs A2C (control) at 3 dpi. b Top 30 DE molecular function and biological process in A5T (infected) vs A5C (control) at 42 dpi. c Top 30 DE molecular function, cellular component and biological process in A6T (infected) vs A6C (control) at 70 dpi
KEGG analysis
KEGG database was used to identify alterations in the biological pathways during F. gigantica infection. A total of 501 transcripts were assigned to 183 KEGG pathways at 3 dpi. At 42 dpi 1,194 transcripts were assigned to 229 KEGG pathways, whereas at 70 dpi 639 transcripts were assigned to 193 KEGG pathways. Top 20 most highly represented pathways in each group are shown in Fig. 5.
Statistics of KEGG pathway enrichment. The x-axis shows the enrichment factor; the y-axis corresponds to KEGG Pathway. The color of the dot represents q value and size of the dot represents the number of DEGs mapped to the reference pathways. a, b and c represent the top 20 statistics of KEGG pathway enrichment for DEGs observed at 3, 42 and 70 dpi, respectively
Transcription factors
AnimalTFDB (http://www.bioguo.org/AnimalTFDB/) was used to identify and classify transcription factors in the buffalo genome. As shown in Fig. 6, there are significant differences among the animal groups. Eighty-two differentially expressed transcription factors were identified in infected livers. For example, Smad 6, which inhibits the transforming growth factor beta (TGF-β) signaling pathway, was upregulated at 3 dpi. Also, B cell lymphoma 6 (Bcl6) was downregulated at 3 dpi, but was upregulated at 42 dpi. The differentially expressed transcription factors were classed into 5 clusters according to their expression patterns: (i) four highly expressed genes at 3 dpi; (ii) 19 highly expressed genes at 42 and 72 dpi; (iii) 43 highly expressed genes at 42 dpi; (iv) 2 highly expressed genes in all groups; and (v) 33 genes clustered into other clusters.
Heatmap of the differentially expressed (DE) transcription factors. a, b and c are differentially expressed transcription factors at 3, 42 and 70 dpi, respectively. The red (up) and green (down) dots represent the significantly differential expressed transcripts; the black represents the transcripts whose expression levels did not reach statistical significance
Discussion
In the present study, we employed RNA-Seq Illumina technology to uncover the hepatic transcriptomic changes of buffaloes infected with F. gigantica. Specifically, we compared the gene expression patterns of the liver of infected and uninfected buffaloes. GO and KEGG enrichment analyses revealed that experimental infection with F. gigantica can influence the expression of genes associated with the host immune response and metabolism. Notably, regulation of some genes indicated potential parasite manipulation to facilitate infection; these included the major histocompatibility complex (MHC) class II (MHC-II) related genes that were repressed, the acute phase protein LBP which was downregulated, modulation of the expression of the transcriptional repressor Bcl6 over the course of infection, upregulation of the transcription factor Smad6, repression of genes involved in the oxidative burst, and finally, modulation of the regenerative response related genes (Brca1 and Blm). The expression patterns of these genes through a time course of 3, 42 and 70 dpi and their relevance to the pathogenesis of F. gigantica infection in buffaloes are discussed in the following sections.
Immune responses
Fasciola hepatica infection in lambs can induce a dominant Th2-biased immune response along with suppression of Th1/Th17 responses [21] and can negatively impact Th1 responses to bystander infections, such as during coinfection with Mycobacterium tuberculosis [41]. Buffaloes, on the other hand, can exhibit a combination of Th1 and Th2 cytokine expression pattern in response to F. gigantica infection [42, 43] or post vaccination with the recombinant fatty acid binding protein (rFABP) and glutathione S-transferase (rGST) protein [44]. Therefore, it is reasonable to expect some differences in the expression patterns of immune response genes that interact with or are stimulated in response to infection with F. gigantica and F. hepatica. Innate immunity is the first line of defense against Fasciola and in the mean time it plays key roles in priming the adaptive immune response [45]. In mammals, antigen processing and presentation are essential for triggering the downstream cellular and/or humoral immune responses [46]. The KEGG results revealed that genes involved in the (MHC-II) pathway were downregulated at 3 dpi, in agreement with others [23], and that genes involved in the MHC-I pathway were upregulated at 42 dpi. Interestingly, at 70 dpi we did not observe any significant alteration in the regulation of MHC-I or MHC-II, suggesting that F. gigantica is capable of evading the host immune system. This unique pattern should be further investigated.
The suppression of the MHC-II related genes during early F. gigantica infection might correlate with a compromised ability of the MHC class II molecules to present processed F. gigantica antigens to CD4(+) T-lymphocytes. Because the stimulation, differentiation, and function of CD4 T cells is central to the development of type I immune responses, any reduction in the interaction between MHC class II molecules and the T cell receptor (TCR) might affect the Th1/Th2 balance, similar to what has been reported in F. hepatica infection [21]. A recent study demonstrated that glycoconjugates from F. hepatica can induce high levels of IL-10 and IL-4, supporting the role of glycans in the polarization of host immune response toward a Th2/regulatory response via induction of IL-10 [25]. Whether the glycosylated molecules of F. gigantica could have the same immune-modulatory effect remains to be investigated.
Lipoppolysaccharide binding protein (LBP), acute phase protein, is produced mainly by hepatocytes [47] and plays an important role in lipopolysaccharide (LPS) signaling and innate immunity [48]. LBP can activate Toll-like receptor 4 (TLR4), which mediates the expression of pro-inflammatory cytokines and other immune response related genes [49]. In our study, the downregulation of LBP at 3 dpi, probably mediated by the parasite fatty acid binding protein [49], suggests that host pro-inflammatory responses may be suppressed during early F. gigantica infection. This result supports previous findings that showed that F. hepatica infection and antigens can suppress Th1 immune responses in vivo [41, 50]. Hence, the reduced hepatic production of the LBP and the subsequent suppression of the pro-inflammatory cytokines might be recognized as mechanistically important for survival of the liver flukes in a hostile host environment. Suppression of inflammation has been reported in sheep liver infected with F. hepatica [28]. Indeed, the importance of mounting strong Th1 immune responses in the protection of the host against challenge infection has been previously demonstrated in vaccination trials in livestock [51]. While the current literature focussed more on the host immune responses to Fasciola has focused on inflammatory responses, our finding suggests that genes that help to keep inflammation in check may also be important in the host response.
Intercellular adhesion molecule-1 (ICAM-1), a member of the immunoglobulin superfamily, is an endothelial- and leukocyte-associated transmembrane protein. ICAM-1 facilitates recruitment of circulating leukocyte (including lymphocytes) to infection/inflamed sites and mediates the interaction between T cells and their target cells [52]. The increase of ICAM-1 expression at 3 dpi is probably beneficial for the adhesion of lymphocytes to the endothelial cells and their migration into the liver, which is a prerequisite of attack of target cells by cytotoxic T lymphocytes. Interestingly, IL-1β was found upregulated at 70 dpi. IL-1β regulates a number of pro-inflammatory genes such as IL-8, a chemokine that attracts neutrophils, and eosinophils, and is involved in antibody-dependent cell-mediated cytotoxicity (ADCC) pathway [53–55], which has a major importance in the defense against Fasciola [26]. At 42 dpi, Itgam, which inhibits Ncf1 and Ncf4, leading to impairment of oxidative burst, was significantly upregulated. This suggests that F. gigantica alters the expression of genes involved in the oxidative burst process in order to avoid killing by nitric oxide (NO), which is considered as a defense mechanism against infection [56]. This finding is consistent with the previous result that NO production and nitric oxide synthase 2 (NOS2) expression are downregulated when monocyte-derived macrophages of human origin were exposed to F. hepatica fatty acid binding protein, known as Fh12 [57]. Our observation is also in agreement with previous studies that reported inability of ovine macrophages to generate NO when incubated with newly excysted juveniles of F. hepatica in vitro [58] and the significant downregulation of NOS2 gene, encoding inducible nitric oxide synthase (iNOS), which converts arginine into citrulline and NO during the acute and chronic stages of ovine F. hepatica infection [21].
We also identified DEGs involved in processes associated with the regulation of Th2 cell differentiation and B-cell activation. We also found, at 3 dpi, upregulation of immunoglobulin variable gene (Ighv1s28, Ig heavy chain Mem5), which inhibits Pro-B cell differentiation to Pre-B1 cell. At 42 dpi, Cd3e, Zap70 and Il-17r genes, which are involved in inhibiting the hematopoietic stem cell (HSC) differentiation, were upregulated. Our study also identified DE transcription factors, such as the transcriptional repressor Bcl6, which is essential for the formation of T-follicular helper (Tfh) cells [59–61], which facilitates T cell-dependent B cell differentiation and antibody responses [62]. Bcl6 was downregulated at 3 dpi, but was upregulated in 42 dpi, suggesting that F. gigantica infection can modulate Bcl6 expression (a traditional regulator of a Th2 response) over the course of infection. This finding is also in agreement with previous work [28] and is consistent with the literature where changing from Th1 to Th2 response occurs as infection establishes, correlating with the development of adaptive B cell response and the generation of Fasciola-specific antibodies within 4 weeks of infection [45]. The transcription factor Smad6 is an inhibitor of TGF-β signaling pathway [63], which plays a key role in fibrosis during F. hepatica infection [64] and can suppress the growth and self-renewal of hepatic progenitor cells [65]. In our study, Smad6 was found upregulated at 3 dpi, suggesting that Smad6 may play a role in controlling fibrosis during the early stage of F. gigantica infection. A previous study reported a similar finding in PBMC isolated from sheep infected with F. hepatica where the expression level of the inhibitory-Smad protein, Smad7, was upregulated, which has been hypothesized to play a role in limiting the fibrosis formation during acute and chronic stage of infection [21].
In contrast, increased levels of PBMC-derived TGF-β1 have been observed in early phases of the infection with F. hepatica in cattle [22]. These findings indicate that molecules of the TGF-β-pathway can potentially exacerbate and ameliorate the liver fibrotic process depending on the stage of the infection, host species and Fasciola species causing the infection.
Metabolic dysregulation
Liver is a very important metabolic and drug clearance organ because changes in the activities or regulation of hepatic drug metabolizing enzymes can alter clearance of chemical compounds. Previous reports indicated that F. hepatica infection can induce alterations in the mitochondrial electron transport chain and the enzymes that are responsible for drug metabolism in the liver [28, 66, 67]. Several hepatic enzymes known to play key roles in the mammalian metabolic and clearance processes, including Flavin monooxygenase (FMO), carboxylesterases (CES), members of cytochrome P450 enzyme, aldehyde dehydrogenase (ALDH), glutathione S- transferase (GST), and paraoxonase (PON), were found to be affected by F. gigantica infection in our study. For example, the expression level of Pon1 in infected liver was 2.2-fold higher compared to the control liver at 3 dpi. At 42 and 70 dpi, infected buffaloes had lower levels of Pon3 (58%) and Ces2 (49%), respectively. At 3 dpi, the mRNA level of Aldh1a1 was 1.6-fold higher than the corresponding control. Other aldehyde dehydrogenases in the infected liver had lower levels [e.g. Aldh3a2 (43%), Aldh1l1 (51%), Aldh4a1 (57%) at 42 dpi and Aldh1l1 (50%) at 70 dpi]. Also, Fmo3 and Fmo5 showed decreased levels (60%) in infected liver samples. The mRNA levels of GST decreased in infected livers at 42 dpi [e.g. Gsta2 (50%) and MGst1 (58%)] and at 70 dpi [e.g. Gsta1 (68%)].
Cytochrome P450 is a very important drug metabolizing enzyme. KEGG analysis revealed that in the cytochrome P450 pathway there are 2 upregulated transcripts (Gstm1 and Gsta3) at 3 dpi, 13 downregulated transcripts (Fmo3, Fmo5, UDP-glucuronosyltransferase 2b4, UDP glucuronosyltransferase 2 family, UDP-glucuronosyltransferase 2C1, Ugt1a6, Ugt2b17, Ugt1a1, Adh5, Gsta2, Gsto1, Mgst1, Maob) at 42 dpi and 4 downregulated transcripts (Ugt2a1, Mgst1, Gsta5, Gsta3) at 70 dpi. The downregulation of cytochrome P450 genes is consistent with previous results [68]. The alterations of these enzymes suggest that infection of buffaloes with F. gigantica infection can modulate drug pharmacokinetics, and this can vary over the course of infection.
Genomic responses
The breast cancer 1 early onset (Brca1), which is a tumor suppressor involved in cellular functions related to cell replication and DNA synthesis was found downregulated at 3 and 42 dpi. Blm, coding blooms syndrome helicase which is a member of the RecQ family of superfamily 2 helicases and plays critical role in the maintenance of genome stability, was found upregulated at 42 dpi. The clinical relevance of the downregulation of Brca1 or upregulation of Blm is still unknown, but it is likely that Bcra1-mediated cell proliferation and Blm-mediated genome integrity play a role in the regenerative response of the liver to heal the damaged tissue caused by F. gigantica infection. A similar observation was reported in sheep livers infected with F. hepatica where genes associated with host cell cycle and mitosis were found significantly upregulated [28].
Bile secretion
Because they live and induce pathological lesions in the bile duct, F. gigantica flukes are expected to interfere with bile synthesis and secretion. Indeed, alteration in the bile production has been reported in F. hepatica infected sheep [69]. The downregulation of the Na+ and D-glucose transfer (Sglt1) gene, expressed at the cholangiocyte apical plasma membrane, at 3 dpi can decrease the apical uptake of glucose from the bile and this could affect the biliary osmolarity. Genes related to the synthesis of carbonic acid (H2CO3) from CO2 and H2O, which facilitates the excretion of bile acid and glutathione (GSH) through kidney were upregulated, potentially causing a reduction in the amount of bile in the biliary duct.
Conclusions
Using RNA sequencing technology, we characterized transcriptome profiles of water buffalo liver during experimental F. gigantica infection. Comparing the infection groups to the mock groups, 496, 880 and 441 significantly DEGs were identified at 3, 42 and 70 dpi, respectively. Infected liver showed alterations in the expression of genes involved in immune responses, hepatic drug metabolism, regenerative response, and bile secretion. Several pathways, such as the MHC antigen processing and presentation, TLR4 signalling, TGF-β signalling, and cytochrome P450 pathway, have been altered by F. gigantica infection. These findings suggest that F. gigantica can modulate host immunity and inflammatory pathways in order to facilitate its survival within the host. A better understanding of the immunopathological mechanisms of F. gigantica infection may provide new preventive and therapeutic strategies for the control of fasciolosis in buffaloes.
Abbreviations
- ADCC:
-
antibody-dependent cell-mediated cytotoxicity
- ALDH:
-
aldehyde dehydrogenase
- AR:
-
anthelmintic resistance
- CES:
-
carboxylesterases
- DEGs:
-
differentially expressed genes
- dpi:
-
days post-infection
- FMO:
-
Flavin monooxygenase
- FPKM:
-
Fragments per kilobase of transcript sequence per millions base pairs sequenced
- GST:
-
glutathione S- transferase
- HSC:
-
hematopoietic stem cell
- ICAM:
-
intercellular adhesion molecule
- LBP:
-
lipopolysaccharide binding protein
- MHC:
-
major histocompatibility complex
- PBMCs:
-
peripheral blood mononuclear cells
- PBS:
-
phosphate buffered saline
- PON:
-
paraoxonase
- RNA-seq:
-
RNA sequencing
- TGF-β:
-
transforming growth factor beta
- TLR4:
-
toll-like receptor 4
References
Mage C, Bourgne H, Toullieu JM, Rondelaud D, Dreyfuss G. Fasciola hepatica and Paramphistomum daubneyi: changes in prevalences of natural infections in cattle and in Lymnaea truncatula from central France over the past 12 years. Vet Res. 2002;33:439–47.
Kuchai JA, Chishti MZ, Zaki MM, Rasool SAM, Ahmad J, Tak H. Some epidemiological aspects of fascioliasis among cattle of Ladakh. Global Vet. 2011;7:342–46.
Spithill TW, Dalton JP. Progress in development of liver fluke vaccines. Parasitol Today. 1998;14:224–8.
Nguyen TG, Le TH, Dao TH, Tran TL, Praet N, Speybroeck N, et al. Bovine fasciolosis in the human fasciolosis hyperendemic Binh Dinh province in Central Vietnam. Acta Trop. 2011;117:19–22.
Machicado C, Machicado JD, Maco V, Terashima A, Marcos LA. Association of Fasciola hepatica infection with liver fibrosis, cirrhosis, and cancer: a systematic review. PLoS Negl Trop Dis. 2016;10, e0004962.
Chen JX, Chen MX, Ai L, Xu XN, Jiao JM, Zhu TJ, et al. An outbreak of human Fascioliasis gigantica in southwest China. PLoS One. 2013;8, e71520.
Inoue K, Kanemasa H, Inoue K, Matsumoto M, Kajita Y, Mitsufuji S, et al. A case of human fasciolosis: discrepancy between egg size and genotype of Fasciola sp. Parasitol Res. 2007;100:665–7.
Ashrafi K, Bargues MD, O'Neill S, Mas-Coma S. Fascioliasis: a worldwide parasitic disease of importance in travel medicine. Travel Med Infect Dis. 2014;12:636–49.
Yadav SC, Sharma RL, Kalicharan A, Mehra UR, Dass RS, Verma AK. Primary experimental infection of riverine buffaloes with Fasciola gigantica. Vet Parasitol. 1999;82:285–96.
Fairweather I, Boray JC. Fasciolicides: efficacy, actions, resistance and its management. Vet J. 1999;158:81–112.
Venturina VM, Alejandro MA, Baltazar CP, Abes NS, Mingala CN. Evidence of Fasciola spp. resistance to albendazole, triclabendazole and bromofenofos in water buffaloes (Bubalus bubalis). Ann Parasitol. 2015;61:283–9.
Koko WS, Galal M, Khalid HS. Fasciolicidal efficacy of Albizia anthelmintica and Balanites aegyptiaca compared with albendazole. J Ethnopharmacol. 2000;71:247–52.
Hossain E, Chandra G, Nandy AP, Gupta JK, Mandal SC. Possible fasciocidal activity of methanol extract of Dregea volubilis leaves. Exp Parasitol. 2013;135:183–7.
Hillyer GV. Induction of immunity in mice to Fasciola hepatica with a Fasciola/Schistosoma cross-reactive defined immunity antigen. Am J Trop Med Hyg. 1985;34:1127–31.
Muro A, Ramajo V, Lopez J, Simon F, Hillyer GV. Fasciola hepatica: vaccination of rabbits with native and recombinant antigens related to fatty acid binding proteins. Vet Parasitol. 1997;69:219–29.
Sexton JL, Milner AR, Panaccio M, Waddington J, Wijffels G, Chandler D, et al. Glutathione S-transferase. Novel vaccine against Fasciola hepatica infection in sheep. J Immunol. 1990;145:3905–10.
Sexton JL, Wilce MC, Colin T, Wijffels GL, Salvatore L, Feil S, et al. Vaccination of sheep against Fasciola hepatica with glutathione S-transferase. Identification and mapping of antibody epitopes on a three-dimensional model of the antigen. J Immunol. 1994;152:1861–72.
Mendes RE, Perez-Ecija RA, Zafra R, Buffoni L, Martinez-Moreno A, Dalton JP, et al. Evaluation of hepatic changes and local and systemic immune responses in goats immunized with recombinant Peroxiredoxin (Prx) and challenged with Fasciola hepatica. Vaccine. 2010;28:2832–40.
Morrison CA, Colin T, Sexton JL, Bowen F, Wicker J, Friedel T, et al. Protection of cattle against Fasciola hepatica infection by vaccination with glutathione S-transferase. Vaccine. 1996;14:1603–12.
Lopez-Aban J, Nogal-Ruiz JJ, Vicente B, Morrondo P, Diez-Banos P, Hillyer GV, et al. The addition of a new immunomodulator with the adjuvant adaptation ADAD system using fatty acid binding proteins increases the protection against Fasciola hepatica. Vet Parasitol. 2008;153:176–81.
Fu Y, Chryssafidis AL, Browne JA, O'Sullivan J, McGettigan PA, Mulcahy G. Transcriptomic study on ovine immune responses to Fasciola hepatica infection. PLoS Negl Trop Dis. 2016;10, e0005015.
Flynn R, Mulcahy G. The roles of IL-10 and TGF-beta in controlling IL-4 and IFN-gamma production during experimental Fasciola hepatica infection. Int J Parasitol. 2008;38:1673–80.
Walsh KP, Brady MT, Finlay CM, Boon L, Mills KH. Infection with a helminth parasite attenuates autoimmunity through TGF-beta-mediated suppression of Th17 and Th1 responses. J Immunol. 2009;183:1577–86.
Hamilton C, Dowling D, Loscher C, Morphew R, Brophy P, O'Neill S. Fasciola hepatica tegumental antigen suppresses dendritic cell maturation and function. Infect Immun. 2009;6:2488–98.
Rodríguez E, Noya V, Cervi L, Chiribao ML, Brossard N, Chiale C, et al. Glycans from Fasciola hepatica modulate the host immune response and TLR-induced maturation of dendritic cells. PLoS Negl Trop Dis. 2015;9(12), e0004234.
Molina EC. Serum interferon-gamma and interleukins-6 and -8 during infection with Fasciola gigantica in cattle and buffaloes. J Vet Sci. 2005;6:135–9.
Chantree P, Phatsara M, Meemon K, Chaichanasak P, Changklungmoa N, Kueakhai P, et al. Vaccine potential of recombinant cathepsin B against Fasciola gigantica. Exp Parasitol. 2013;135(1):102–9.
Alvarez Rojas CA, Ansell BR, Hall RS, Gasser RB, Young ND, Jex AR, et al. Transcriptional analysis identifies key genes involved in metabolism, fibrosis/tissue repair and the immune response against Fasciola hepatica in sheep liver. Parasit Vectors. 2015;8:124.
Alvarez Rojas CA, Scheerlinck JP, Ansell BR, Hall RS, Gasser RB, Jex AR. Time-course study of the transcriptome of peripheral blood mononuclear cells (PBMCs) from sheep infected with Fasciola hepatica. PLoS One. 2016;11, e0159194.
Hacariz O, Akgun M, Kavak P, Yuksel B, Sagiroglu MS. Comparative transcriptome profiling approach to glean virulence and immunomodulation-related genes of Fasciola hepatica. BMC Genomics. 2015;16:366.
Molina EC, Skerratt LF. Cellular and humoral responses in liver of cattle and buffaloes infected with a single dose of Fasciola gigantica. Vet Parasitol. 2005;131:157–63.
Molina EC, Gonzaga EA, Sinolinding EO, Lumbao LA, Peralta AA, Barraca AP. Differences in susceptibility between cattle and swamp buffaloes to infection with Fasciola gigantica. Trop Anim Health Prod. 2005;37:611–6.
Phalee A, Wongsawad C, Rojanapaibul A, Chai JY. Experimental life history and biological characteristics of Fasciola gigantica (Digenea: Fasciolidae). Korean J Parasitol. 2015;53:59–64.
Chauvin A, Bouvet G, Boulard C. Humoral and cellular immune responses to Fasciola hepatica experimental primary and secondary infection in sheep. Int J Parasitol. 1995;25:1227–41.
Sipka S, Bruckner G. The immunomodulatory role of bile acids. Int Arch Allergy Immunol. 2014;165:1–8.
Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14.
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4.
Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–93.
Flynn RJ, Mannion C, Golden O, Hacariz O, Mulcahy G. Experimental Fasciola hepatica infection alters responses to tests used for diagnosis of bovine tuberculosis. Infect Immun. 2007;75:1373–81.
Changklungmoa N, Phoinok N, Yencham C, Sobhon P, Kueakhai P. Vaccine potential of recombinant cathepsinL1G against Fasciola gigantica in mice. Vet Parasitol. 2016;226:124–31.
Kumar N, Raina OK, Nagar G, Prakash V, Jacob SS. Th1 and Th2 cytokine gene expression in primary infection and vaccination against Fasciola gigantica in buffaloes by real-time PCR. Parasitol Res. 2013;112:3561–8.
Kumar N, Anju V, Gaurav N, Chandra D, Samanta S, Gupta SC, et al. Vaccination of buffaloes with Fasciola gigantica recombinant glutathione S-transferase and fatty acid binding protein. Parasitol Res. 2012;110:419–26.
Flynn RJ, Mulcahy G, Elsheikha HM. Coordinating innate and adaptive immunity in Fasciola hepatica infection: implications for control. Vet Parasitol. 2010;169:235–40.
Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–18.
Ramadori G, Buschenfelde KH M z, Tobias PS, Mathison JC, Ulevitch RJ. Biosynthesis of lipopolysaccharide-binding protein in rabbit hepatocytes. Pathobiology. 1990;58:89–94.
Finberg RW, Re F, Popova L, Golenbock DT, Kurt-Jones EA. Cell activation by Toll-like receptors: role of LBP and CD14. J Endotoxin Res. 2004;10:413–8.
Martin I, Caban-Hernandez K, Figueroa-Santiago O, Espino AM. Fasciola hepatica fatty acid binding protein inhibits TLR4 activation and suppresses the inflammatory cytokines induced by lipopolysaccharide in vitro and in vivo. J Immunol. 2015;194:3924–36.
Brady MT, O'Neill SM, Dalton JP, Mills KHG. Fasciola hepatica suppresses a protective Th1 response against Bordetella pertussis. Infect Immun. 1999;67:5372–8.
Mulcahy G, O'Connor F, McGonigle S, Dowd A, Clery DG, Andrews SJ, Dalton JP. Correlation of specific antibody titre and avidity with protection in cattle immunized against Fasciola hepatica. Vaccine. 1998;16:932–9.
Imhof BA, Dunon D. Leukocyte migration and adhesion. Adv Immunol. 1995;58:345–416.
Borish LC, Steinke JW. 2. Cytokines and chemokines. J Allergy Clin Immunol. 2003;111:S460–75.
Mitchell GB, Albright BN, Caswell JL. Effect of interleukin-8 and granulocyte colony-stimulating factor on priming and activation of bovine neutrophils. Infect Immun. 2003;71:1643–9.
Reali E, Spisani S, Gavioli R, Lanza F, Moretti S, Traniello S. IL-8 enhances antibody-dependent cellular cytotoxicity in human neutrophils. Immunol Cell Biol. 1995;73:234–8.
Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2(10):907–16.
Figueroa-Santiago O, Espino AM. Fasciola hepatica fatty acid binding protein induces the alternative activation of human macrophages. Infect Immun. 2014;82:5005–12.
Piedrafita D, Parsons JC, Sandeman RM, Wood PR, Estuningsih SE, Partoutomo S, et al. Antibody-dependent cell-mediated cytotoxicity to newly excysted juvenile Fasciola hepatica in vitro is mediated by reactive nitrogen intermediates. Parasite Immunol. 2001;23(9):473–82.
Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, et al. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188:3734–44.
Nakayamada S, Poholek AC, Lu KT, Takahashi H, Kato M, Iwata S, et al. Type I Interferon induces binding of STAT1 to Bcl6: Divergent Roles of STAT-family transcription factors in the TFH cell genetic program. J Immunol. 2014;192:2156–66.
Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, et al. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS One. 2011;6, e17739.
Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, et al. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–52.
Macias MJ, Martin-Malpartida P, Massague J. Structural determinants of SMAD function in TGF-β signaling. Trends Biochem Sci. 2015;40:296–308.
Hacariz O, Sayers G, Flynn RJ, Lejeune A, Mulcahy G. IL-10 and TGF-beta1 are associated with variations in fluke burdens following experimental fasciolosis in sheep. Parasite immunol. 2009;31(10):613–22.
Ding ZY, Liang HF, Jin GN, Chen WX, Wang W, Datta PK, et al. Smad6 suppresses the growth and self-renewal of hepatic progenitor cells. J Cell Physiol. 2014;229:651–60.
Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–41.
Tufenkji AE, Alvinerie M, Larrieu G, Houin G, Galtier P. Pharmacokinetics of ampicillin and pentobarbital in the course of subclinical fascioliasis in sheep. Res Vet Sci. 1991;50:75–80.
Giorgi M, Salvatori AP, Soldani G, Giusiani M, Longo V, Gervasi PG, et al. Pharmacokinetics and microsomal oxidation of praziquantel and its effects on the P450 system in three-month-old lambs infested by Fasciola hepatica. J Vet Pharmacol Ther. 2001;24:251–9.
Lopez P, Gonzalez P, Tunon MJ, Gonzalez-Gallego J. The effects of experimental fasciolosis on bilirubin metabolism in the rat. Exp Parasitol. 1994;78:386–93.
Acknowledgements
The authors would like to thank Novogene Bioinformatics Technology Co., Ltd (Beijing, China) for performing the sequencing and preliminary data analysis.
Funding
Project financial support was provided by the National Key Basic Research Program (973 Program) of China (Grant No. 2015CB150300), the Fundamental Research Funds for Central Research Institutes of Public Interests (Grant No. 1610312017022) and the Fundamental Research Funds of Chinese Academy of Agricultural Sciences (Grant No. Y2016JC05).
Availability of data and materials
The datasets supporting the findings of this article are included within the article. The RNA-seq raw data are available in the NCBI SRA repository under accession number PRJNA341921.
Authors’ contributions
XQZ and AJG conceived and designed the study, and critically revised the manuscript. FKZ and XXZ performed the experiment, analyzed the transcriptomic data and drafted the manuscript. HME and JJH helped in data analysis and manuscript revision. ZAS, WBZ, JGM and WYH helped in the implementation of the study. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
The study design was reviewed and approved by the Animal Ethics Committee of Lanzhou Veterinary Study Institute, Chinese Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1:
Figure S1. Liver of an infected buffalo showing adult Fasciola gigantica fluke in situ. (TIF 1827 kb)
Additional file 2:
Figure S2. Adult Fasciola gigantica fluke isolated from the liver of an infected buffalo. (TIF 6665 kb)
Additional file 3:
Table S1. Summary of all data obtained from all samples in the present study. (DOC 46 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, FK., Zhang, XX., Elsheikha, H.M. et al. Transcriptomic responses of water buffalo liver to infection with the digenetic fluke Fasciola gigantica . Parasites Vectors 10, 56 (2017). https://doi.org/10.1186/s13071-017-1990-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-017-1990-2