Abstract
Background
There is growing evidence that the widespread use of Long-Lasting Insecticidal Nets (LLINs) is prompting malaria vectors to shift their biting towards times and places where people are not protected, such as earlier in the evening and/or outdoors. It is uncertain whether these behavioural shifts are due to phenotypic plasticity and/or ecological changes within vector communities that favour more exophilic species, or involve genetic factors within vector species to limit their contact with LLINs. Possibly variation in the time and location of mosquito biting has a genetic basis, but as yet this phenomenon has received little investigation. Here we used a candidate gene approach to investigate whether polymorphisms in selected circadian clock genes could explain variation in the time and location of feeding (indoors versus outside) within a natural population of the major African malaria vector Anopheles arabiensis.
Methods
Host-seeking An. arabiensis were collected from two villages (Lupiro and Sagamaganga) in Tanzania by Human Landing Catch (HLC) technique. Mosquitoes were classified into phenotypes of “early” (7 pm–10 pm) or “late” biting (4 am –7 am), and host-seeking indoors or outdoors. In these samples we genotyped 34 coding SNPs in 8 clock genes (PER, TIM, CLK, CYC, PDP1, VRI, CRY1, and CRY2), and tested for associations between these SNPs and biting phenotypes. SNPs in 8 mitochondrial genes (ATP6, ATP8, COX1, COX2, COX3, ND3, ND5 and CYTB) were also genotyped to test population subdivision within An. arabiensis.
Results
The candidate clock genes exhibited polymorphism within An. arabiensis, but it was unrelated to variation in the timing and location of their biting activity. However, there was evidence of strong genetic structure within An. arabiensis populations in association with the TIM, which was unrelated to geographic distance. Substructure within An. arabiensis was also detected using mitochondrial markers.
Conclusions
The variable timing and location of biting in An. arabiensis could not be linked to candidate clock genes that are known to influence behaviour in other Diptera. This finding does not rule out the possibility of a genetic basis to biting behaviour in this malaria vector, but suggests these are complex phenotypes that require more intensive ecological, neuronal and genomic analyses to understand.
Similar content being viewed by others

Background
In Africa, the prominent malaria vector species include Anopheles gambiae Giles sensu stricto (s.s.); Anopheles arabiensis Patton and An. coluzzii Coetzee & Wilkerson are members of the An. gambiae sensu lato (s.l.) species complex. Females of these species require vertebrate blood to develop their eggs, and regularly feed upon humans in the wild [1]. The stereotypical pattern of host-seeking in these vector species was described in early work by Gillies [2], and is characterised by the onset of a daily period of host-seeking which begins after dusk and increases to a peak around midnight, with 60–80 % of bites estimated to occur between 9 pm and 3 am [3]. This host-seeking activity coincides with the period when most people are indoors and asleep [1, 4]. This pattern of behaviour underlies the success of control measures like Long-Lasting Insecticidal Nets (LLINs) by selectively protecting people when they are asleep at night indoors [5].
However, host-seeking behaviour varies between species within the An. gambiae complex. For example, An. gambiae (s.s.) is highly endophagic (preference to feed indoors) and anthropophagic (preference for biting humans) [6] and feeds predominantly between 9 pm–3 am [7]. Its sibling species An. arabiensis can be relatively exophagic and zoophagic (feeding on cattle as well as humans) [6, 8–10], and can spread its biting over a wider period of the night with less distinct peaks in activity [11]. This provides opportunities for vectors like An. arabiensis to evade control using LLINs [12].
Within the An. gambiae (s.l.) species complex, there have been reports of shifts in their behaviours such as increased tendency to feed outdoors [13, 14], to bite earlier or later in the night [15], and reduced anthropophagy [16] in the presence of vector control measures [17–22]. These behavioural shifts have been associated with changes in the species composition towards vector species with more exophilic behaviour in East Africa. Whilst An. gambiae s.s. was the most abundant member of the An. gambiae (s.l.) species complex [23], in some places in Africa this species has significantly declined in association with bed-net use and now An. arabiensis constitutes >95 % of the complex [24, 25]. Whilst environmental contributions to malaria vector feeding behaviour have been documented (e.g. increased zoophily and outdoor resting in the presence of livestock [26]), the influence of mosquito genetic factors on their host-seeking activity is poorly understood. Given that the capacity for mosquitoes to rapidly adapt their feeding activity to avoid contact with LLINs [27], there is a need to investigate if this phenomenon has genetic basis and therefore if it can be subjected to selection imposed by mosquito control measures.
The location in which malaria vectors prefer to bite (e.g. indoors or outdoors) has some genetic basis [28]. For example, a chromosome inversion 2Rbc is associated with outdoor biting and resting behaviour in An. arabiensis [29]. Furthermore, the 2Ra and 3Ra inversions are associated with endophagy in An. funestus [30]. Whilst these studies support the hypothesis that variation in mosquito feeding behaviour is influenced by genetic factors, their use of coarse genetic units such as chromosomal inversions makes it difficult to establish a causal relationship with specific functional genes. The use of more modern, fine-scale genetic analysis approaches based on single nucleotide polymorphisms will enable a much more powerful investigation of phenotype-genotype associations with greater likelihood of identifying the specific genes that influence behaviour [31, 32].
Circadian clock genes are obvious targets for investigation of genes responsible for variation in the timing of daily activity patterns, specifically feeding rhythms in mosquitoes. In Drosophila, Period (PER) and Timeless (TIM) have been identified as key clock genes that encode critical components of the molecular oscillator that drives circadian rhythms. PER and TIM proteins are transcriptional repressors that regulate their own expression through negative feedback mechanisms by suppressing the activity of CLOCK [CLK] and CYCLE [CYC], two activators of PER and TIM transcription [33]). Vrille (VRI) and par domain protein 1 (PDP1) encodes additional transcription factors with opposing function to control the expression level of CLK [34, 35]. Doubletime (DBT) is also an important clock gene in Drosophila, where it encodes a clock kinase that influences circadian rhythmicity through phosphorylation of PER protein, thereby setting the speed of the clock [33]. Additional clock genes include Cryptochrome 1 (CRY1), which encodes a flavin binding photoreceptor found ubiquitously in mammals, insects and plants [36], and regulate circadian activities by facilitating the degradation of TIM protein via the proteasomal pathway in the presence of light [37]. On the other hand, Crytochrome 2 (CRY2), which is only present in some insect species and in mammals, has been shown to encode a transcription factor that replaces PER as a repressor of CLK or BMAL1 protein activity [38]. These genes can therefore serve as a useful starting point for investigating the genetic basis of polymorphisms in the feeding behaviour of malaria vectors.
So far, only a few studies have investigated clock genes in An. gambiae (s.l.) and their potential association with diel activity such as blood feeding. Previous studies applied transcriptomic approaches to study gene expression patterns [39–41], and established a genome-wide profiling of circadian gene expression in An. gambiae (s.s.) [42]. However, there has been little investigation of the link between mosquito feeding behaviours and single nucleotide polymorphisms (SNPs) in clock genes occurring in natural populations.
Here, we hypothesise that mutations in clock genes may explain variation in the host-seeking times of African malaria vectors. We used SNPs to test for associations between SNPs in exons of 8 clock genes and the times at which An. arabiensis were caught host-seeking within a natural population in Tanzania. We tested whether any observed associations were consistent within mosquitoes attempting to feed indoors and outdoors. We acknowledge that time of feeding can be influenced by many environmental factors such as distance from host, wind direction, wind speed and availability of other hosts. While these variables were somewhat mitigated by conducting all collections within two days per village and in multiple villages, we expect we would be able to detect significant associations if genetic contribution toward the time of feeding phenotype is relatively strong (effect size of 0.6 or greater; see Methods section below for power analysis).
Methods
Study sites
Anopheles arabiensis were collected in the villages of Lupiro (−08.38′S, 36.67′E) and Sagamaganga (−08.07′S; 36.80′E), which are situated about 40 km apart in the Kilombero Valley of Tanzania (Fig. 1). This area has endemic, year-round malaria transmission [43–45] that peaks in the rainy seasons occurring from November to January and from March to May. Residents of Lupiro and Sagamaganga villages are primarily subsistence rice cultivators. Pastoralism is also common in Sagamaganga with many local residents keeping cattle. Both villages have experienced a significant change in the species composition of malaria vectors within recent years as LLINS have reached near 100 % coverage levels.
Behavioural phenotype selection and mosquito sample collection
Mosquitoes were collected at paired indoor and outdoor trapping stations in three experimental huts per night. Experiments were done between February and March 2013. Mosquitoes were collected by Human Landing Catch (HLC) technique as they attempted to feed on people. These collections were conducted by a volunteer who sat on a chair with his legs exposed from foot to knee. Using a mouth aspirator, the volunteer sucked up mosquitoes as soon as they landed on his exposed legs. Collections were made from 19:00 to 07:00 the next morning, with volunteers actively catching for 45 min in each hour then using the remaining period to rest. Collectors were swapped between different trap types and location of collection to minimise bias that might arise due to differential collectors’ attractiveness to mosquitoes. Captured mosquitoes were placed into holding cups, grouped for each hour of collection. Those morphologically identified as being An. gambiae (s.l.) were stored in 80 % ethanol to preserve DNA for downstream molecular assays. Anopheles gambiae (s.l.) specimens were classified into one of four categories based on the time and location they were caught host-seeking: (1) indoor early feeding, (2) indoor late feeding, (3) outdoor early feeding, and (4) outdoor late feeding. Early feeding was defined as mosquitoes collected whilst host-seeking in the early hours of the night (19:00–22:00), while late feeding mosquitoes were those collected whilst host-seeking between 04:00 and 07:00.
Species identification and SNP discovery analysis
Genomic DNA was extracted from mosquito samples using DNeasy extraction kits (QIAgen, Valencia, CA, USA). First, PCR analysis was performed on specimens to identify their species within the An. gambiae (s.l.) complex according to Scott’s method [46]. All successfully amplified samples were An. arabiensis. This resulted in a final sample size of between 91 and 96 An. arabiensis for each phenotype and village combination for genetic analysis. If the phenotype has any genetic basis, we expect to detect association between time of feeding behaviour and genotypes of which effect size is 0.6 or greater with 80 % power given our sample size of N > 90 per group. We used pwr library in R software package for power analysis.
Eight clock genes originally identified from the An. gambiae (s.s.) genome [42] were selected for SNP discovery using conventional Sanger sequencing: Period (PER), Timeless (TIM), Clock (CLK), Cycle (CYC), Pdp1 (PDP1), Vrille (VRI), Cryptochrome1 (CRY1), and Cryptochrome2 (CRY2). These genes were selected on the basis of their known association with circadian rhythmic behaviours or function within the molecular oscillator in other insect taxa including An. gambiae (s.s.) [42, 47, 48]. A series of primers were designed for each gene fragment using Primer3 online tools (http://frodo.wi.mit.edu/primer3/). The identity, specific loci sequenced and primer sequences used for all 8 candidate genes are presented in Table 1.
Initially, 8 An. arabiensis samples from each of the four feeding phenotypes at both sites were sequenced for SNP discovery (n = 64). From the 8 genes selected, a total of 34 loci were picked for sequencing. These loci were selected from conservative regions of DNA and included synonymous and non-synonymous mutations that have a codon frequency change of 2 or greater, as is the standard approach to identify mutations that are most likely to influence protein function [49]. The identity of these genes as obtained from the An. gambiae s.s. genome (https://www.vectorbase.org/faqs ) and their chromosomal locations are shown in Table 2.
All 8 clock genes were PCR amplified in a reaction which included 0.2 X Q solution (QIAgen) 1X buffer (Sigma-Aldrich), 1 mM MgCl, 0.4 mM DNTP, 0.1 mM forward primer, 0.1 mM reverse primer, 1 Unit of HotstarTaq Plus DNA polymerase (QIAgen), ~8 μg/μl DNA and pure water. Successful amplification of PCR products was verified using QIAxcel ScreenGel (QIAgen) software version 1.2. The concentration of DNA was measured by spectrophotometry using NanoDrop 1000 V3.7 (Thermo Fisher Scientific Inc.). Ten microliters of the amplified DNA products were purified using DNASap purification kit and sent for conventional post-PCR Sanger sequencing at a DNA sequencing facility at the University of California Davis. Sanger sequencing was used on a subset of the 64 samples used for the purpose of SNP discovery. Each gene fragment was sequenced in both forward and reverse directions. Sequences were checked for quality control using Geneious software version 6.1 [50], in which manual alignment of forward and reverse strands was conducted. Poor quality sequences were trimmed, and alignment of reads to reference sequences was made.
SNP genotyping assay and statistical analysis
The Typer Assay Designer software (Agena Biosciences, San Diego, CA) was used to devise a multiplex SNP genotype assay to allow screening of SNP polymorphisms within the samples. These samples were genotyped using the Agena Biosciences MassARRAY iPLEX platform for the full set of 34 loci derived from the 8 clock genes. Negative controls were run for each plate of samples genotyped. A signal to noise ratio of 3 or above the background level was used to call genotypes. The TyperAnalyzer Application (Agena Biosciences, version 4.0.24.71) was used to score genotypes across all 34 loci. The population genetics software DnaSP v.5 [51] was used to identify haplotype sequences from the iPLEX using a phase algorithm to score SNP density, calculate Tajima D statistics, the number of nucleotide substitutions per phenotype based on direct sequencing results, and the number of shared mutations between groups.
The Arlequin software version 3.5 [52] was used to test departure from Hardy-Weinberg equilibrium (HWE) within each locus. The STRUCTURE software [53] was used to conduct clustering analysis to assign populations or individuals into their membership groups based on the feeding behavioural phenotypes (i.e. time and location of feeding). STRUCTURE was run through a burn-in period of 50,000 followed by 50,000 replications. Results from STRUCTURE were uploaded into STRUCTURE Harvester [54] to assess the ∆K statistic according to Evanno [55] in order to select the number of distinct genetic clusters (K) represented within samples. To determine how the TIM gene influenced the clustering of samples based on the STRUCTURE, separate STRUCTURE analyses were performed in four groups: the first group included all 8 genes (34 loci), the second group excluded TIM (30 loci included), the third group included TIM alone (4 loci) and the fourth group involved analysis done on the non-synonymous SNPs alone to sort out the possible noise effect of the synonymous SNPs on the clustering analysis. In all STRUCTURE analyses, eight populations (K = 8) were assumed a priori. Finally, IndQsort (http://grassi2.ucdavis.edu/~yoosook/Scripts/indQsort/) was used to reorder individuals according to their membership coefficients. Visualization of population clustering was obtained using the Distruct software [56].
Further confirmation of the effect of TIM in the population clustering was assessed by Fisher Exact tests [57] performed by comparing the distribution of the frequencies of each of the 34 loci between the 2 clusters generated by STRUCTURE. To avoid the possibility of obtaining false positive results due to multiple comparisons, a Bonferroni correction [58] was done by dividing the P-values by the number of comparisons performed. Additional confirmation of the genetic clustering analysis was done using Principal Component Analysis (PCoA) implemented in GenALEx [59] and available as a plug-in in Excel. PCoA was done for all 34 loci in two steps. Step 1 involved a pull of all 8 phenotypes, while in step 2 each of the 8 phenotypes was analysed separately.
A further study was conducted on published genome sequences of An. arabiensis [60] from samples that overlap with our study area (Minepa, Lupiro and Sagamaganga) including an out-group population from Cameroon in West Africa. These genome sequences were mapped to An. gambiae mitochondrial sequence ([55]; GenBank ID: NC_002084) using BWA-MEM [61]. Mitochondrial sequences for 24 individual sequences were aligned using Geneious program v.6.1.4. Jukes-Cantor Genetic distance model with bootstrap was used to draw phylogenetic tree.
The 8 SNPs from 8 mitochondrial genes (ATP6, ATP8, COX1, COX2, COX3, ND3, ND5 and CYTB) were selected for SNP genotyping 87 An. arabiensis samples from 4 feeding phenotypes (early feeding, late feeding, indoor feeding and outdoor feeding) collected in Sagamaganga and Lupiro. Collection of these samples was done in households using HLC technique. Analysis of these samples followed the same procedures as outlined above. The SNP information including flanking sequences and primer sequences used for iPLEX SNP genotyping are provided in Additional file 1: Table S4. This additional analysis aimed at establishing support for possible population substructure among An. arabiensis in the Kilombero valley.
Ethics statement
The National Institute for Medical Research (NIMR) provided ethical clearance certificate number NIMR/HQ/R.8a/Vol. IX/801 that allowed this research to be conducted. Informed consent forms were prepared by Ifakara Health Institute (IHI) and submitted for approval to the NIMR. Before setting up experiments, household members were informed of the procedures of the experiments and had to read the informed consent forms before participating in the experiments. Willing household owners signed the forms to allow experiments to be done in and around their houses. Volunteers were informed of the procedures to observe during sample collection including risks involved. Participating volunteers were given malaria prophylaxis Malarone (250 mg atovaquone and 100 mg proguanil hydrochloride, GlaxoSmithKline) before and during the experiments to prevent infection.
Results
All 762 An. gambiae s.l. from Lupiro and Sagamaganga villages analysed were An. arabiensis. We discovered an average of one SNP in every 46.8 ± 34.5 base-pairs in 8 clock genes, with CYC and VRI having the highest and the lowest densities of 1 SNP per every 10 and 125 bp respectively. This was comparable to the overall SNP density previously reported for An. arabiensis (1 every 47 bp [62]).
iPLEX SNP genotyping
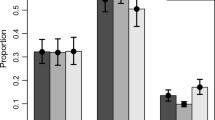
Twenty-one of the 34 loci genotyped had synonymous mutations, while the remaining 13 loci had non-synonymous mutations (Table 3). SNPs in TIM had the highest number of synonymous mutations (ranging from 10 to 15, Table 4) in all the 4 phenotypes. Non-synonymous mutations in TIM ranged from 1 to 2, which was notably lower than those reported in the two genes with the highest rates of non-synonymous mutations (e.g. CRY2 and CYC (10–21, Table 4). The remaining genes had low to moderate numbers of synonymous and non-synonymous mutations ranging from 0 to 7 in both cases. No fixed polymorphisms were detected.
There was no evidence of genetic distance between An. arabiensis samples from different feeding locations (indoor vs outdoor, FST <0.001), feeding times (early vs late, FST <0.001), or geographical locations (Sagamaganga vs Lupiro, FST <0.001). None of the Tajima’s D values were significant (Table 3), indicating the SNPs are evolving neutrally with no evidence of selection, demographic expansion or contraction. All SNPs were in Hardy-Weinberg equilibrium, suggesting an absence of selection on these candidate genes within the populations. STRUCTURE assigned individuals into two distinct groups based on observed patterns of genetic variation (Fig. 2). However, this substructure was not associated with feeding behaviour or site of collection. Analysis of the membership coefficients of cluster 1 and 2 revealed that the frequencies of SNPs in TIM were most divergent between two clusters (Table 4). Fisher Exact tests on the frequencies of all the 34 loci between cluster 1 and 2 showed that there was no difference in the frequency of 30 loci distributed between cluster 1 and 2, while the 4 loci belonging to TIM showed strong significant difference (P < 0.0001) in their distribution among cluster 1 and 2 following Bonferroni correction (Table 5) implying that TIM strongly drives the binary clustering observed in the samples.
STRUCTURE clustering results with parameter K = 2. All 34 loci icluded in (a), and in (b) only the 4 TIM loci included. LEI=Lupiro early indoors, LLI=Lupiro late indoors, SEI=Sagamaganga early indoors, SLI=Sagamaganga late indoors, LEO=Lupiro early outdoors, LLO=Lupiro late outdoors, SEO=Sagamaganga early outdoors, SLO=Sagamaganga late outdoors. There appears to be two population sub-divisions (blue and yellow) across samples with different feeding behaviours
Further analysis of the clustering results from STRUCTURE was carried with the aim of determining the effect of TIM on the population structure of An. arabiensis in the study site. Analysis of clustering results from STRUCTURE performed on all 34 loci including the 4 loci from TIM gave a moderate support for 2 clusters irrespective of the feeding phenotypes and origin of the sample (Additional file 2: Figure S1A). Support for K = 2 was stronger when loci from TIM alone were analysed separately (Additional file 2: Figure S1B) suggesting that TIM played a key role in the binary structuring of An. arabiensis.
Principal Component Analysis (PCoA) results of all 8 phenotypes combined showed that there was no population sub-division based on the feeding phenotypes (Fig. 3), confirming the same results obtained from the STRUCTURE analysis. PCoA revealed two sub-populations more pronounced in samples from Lupiro compared to those from Sagamaganga (Fig. 3). This suggests presence of two sub-populations irrespective of the feeding behaviour of mosquitoes in Lupiro and Sagamanga villages. A geographical association in the sub-structuring of An. arabiensis is yet to be established.
Principal Component Analysis based on the genetic distances generated by STRUCTURE at K = 2. All 8 phenotypes were combined in one analysis and separate analysis for each of the 8 feeding phenotypes. Coordinate 1 and 2 represent the first and second principal components, respectively. PCoA analysis included a total of 730 samples from 8 feeding phenotypes from Lupiro and Sagamaganga: LEI (Lupiro early indoors), LEO (Lupiro early outdoors), LLI (Lupiro late indoors), LLO (Lupiro late outdoors), SEI (Sagamaganga early indoors), SEO (Sagamaganga early outdoors), SLI (Sagamaganga late indoors), SLO (Sagamaganga late outdoors). All 34 loci were included in the PCoA analysis
Further support for population sub-structuring in the Kilombero valley
We further investigated the population structure within An. arabiensis in our study site using the published genome sequence data of An. arabiensis, which overlap with our study area of Lupiro and Sagamaganga [60]. We identified fixed nucleotide differences between Minepa and Sagamaganga lineages in 7 out of 8 SNPs from 8 mitochondrial genes (Additional file 3: Table S2). We also found two major genetic clusters in samples from the Kilombero valley and those from Cameroon based on the mitochondrial sequences (Fig. 4). Analysis revealed samples from the Kilombero valley to be subdivided into two major lineages. The two major lineages are Lineage 1 that includes samples from Sagamaganga, Minepa and Lupiro, and Lineage 2 that includes samples from Minepa and Lupiro (Fig. 4). Samples from Cameroon and one from Minepa clustered out in a separate group (Lineage 3, Fig. 4). These results indicate that Tanzanian population of An. arabiensis is subdivided, as was suggested in [63]. Subdivision within Tanzania appears to be somewhat geographically related. However, from the 87 samples from four feeding phenotypes from Lupiro and Sagamaganga that were genotyped, two lineages were found in both sites regardless of the phenotype group (Additional file 4: Table S3). The reproductive isolation between two lineages has yet to be determined. Overall the genetic clusters based on mitochondrial sequence supports that there are at least two populations of An. arabiensis in our study site that are not related geographically nor according to biting time differences.
Unrooted tree based on Jukes-Cantor Genetic distance model with bootstrap using complete mitochondrial sequences extracted from [60]. Three distinct groups were identified: Sagamaganga group which included samples from Sagamaganga and Lupiro, Minepa group with samples from Minepa and Lupiro and the Cameroon group
Discussion
This study investigated genetic diversity within An. arabiensis with the aim of assessing whether variation in a set of eight clock genes, which critical components of the molecular oscillator controlling rhythmic behaviour, could explain differences in the time and location (indoors versus outdoor) of their host-seeking behaviour. In total, 34 loci incorporating 313 polymorphic sites were assayed. Based on this subset of genes, there was no evidence that the feeding behavioural phenotypes (early vs late and indoor vs outdoor feeding) of An. arabiensis clustered as distinct genetic subpopulations.
There may be several reasons for this lack of association. First, it should be recognised that the molecular oscillator controlled by clock genes is still poorly understood in haematophagous insects, in contrast to Drosophila species [47]. Whilst the candidate clock genes used here were drawn from studies in Drosophila and have been shown to have time-dependent expression in An. gambiae (s.s.) [41], their mode of action and daily expression patterns in relation to rhythmic activities in An. arabiensis have not yet been confirmed. Additionally, lack of correlation of SNPs in core clock genes and timing of biting behaviour highlights the possibility that alterations in feeding rhythms could be due to changes in the neuronal network properties and/or activity that connects the molecular oscillator to output mechanisms that control feeding. Finally, failure to link genetic mutations to feeding behaviour phenotypes in An. arabiensis could be due to methodological issues, including the usage of too few markers, as linkage disequilibrium in An. arabiensis has been shown to breakdown within 200 bp [60]. Other reasons could be, use of inappropriate markers, imprecise classification of phenotypes and/or the fact that extensive phenotypic plasticity in feeding behaviour is possible within one genotype [64]. Use of individuals from the extreme of the time continuum, e.g. those that bite during the day and those that bite late at night, could increase chances of finding association between feeding and genetic structure. However, this is practically impossible as An. arabiensis are not known to feed during the day, making it possible only to sample mosquitoes which feed during the night.
A potential limitation of this study, which may have reduced the ability to identify clear phenotype-genotype associations, was that the biting time phenotypes were quite coarsely and perhaps imprecisely defined. Selection of phenotypes in this study was based on broad categorisation into “early” vs “late” feeding groups, with each period spanning 3 h of collection. Though such categorisation was based on evidence from previous studies which showed that some mosquitoes prefer to feed early at dawn while some feed late at night [15, 65–67], it is not evident that the two phenotypes represent two genetically different groups or just the same group feeding at two different periods of the night. Further studies involving finer scale timing of the feeding behaviour, for example within an hourly interval could be tested in the future. Another potential imprecision is that the time at which mosquitoes were collected may not necessarily have reflected the time at which they initiated their host-seeking. For example, those caught during the late period may actually have begun feeding during the early period of the night, but been unsuccessful in locating a host. Other factors such as distance from host, wind direction, wind speed, and availability of other hosts were not discussed, because all the samples used in this analysis were collected within two days period in each location. The availability of other hosts was different between two locations, Lupiro and Sagamaganga, but we did not detect any difference in feeding time profile between two locations. It would be difficult to assess this under natural conditions, but more detailed investigation of a small number of mosquitoes under lab or semi-field settings may be viable.
Finally, although the clock genes investigated here were not linked with feeding behaviours, variation noted here may be associated with other behaviours which influence gene flow. Specifically the clustering of mosquito population into two groups by the TIM in this study may indicate presence of two coexisting populations that have mating incompatibility possibly through temporal and cytological incompatibility.
Whilst behavioural phenotypes showed no genetic basis here, there was evidence of strong genetic clustering within An. arabiensis samples in association with TIM. The association with TIM was so strong that the same pattern of genetic structure was predicted from this gene alone as with all eight clock genes combined (Additional file 2: Figure S1). Studies have shown that TIM is involved in regulating circadian rhythms, which may influence the time of feeding [39, 68]. TIM has also been found to regulate autophagy and diapause [68]. Furthermore, markers based on TIM alone have been used to identify population structure in Anopheles cruzii Dyar & Knab [69], and in Anopheles triannulatus s.l. (Neiva & Pinto) [70]. Additionally, TIM has also shown variation in expression levels across different times of day in the pitcher-plant mosquito Wyeomyia smithii (Coquillett) [71]. The link between TIM and population structure in these studies suggests that this gene may also be playing a role in population structure of An. arabiensis.
While it may not be surprising to find samples from different feeding phenotypes and from the 40 km apart villages of Lupiro and Sagamaganga showing no genetic clustering, it is however interesting for samples within the villages to cluster based on TIM. Studies involving other insects have shown population clustering based on TIM across geographical locations. For example, in a study of frequencies of allele (ls-tim), which is one of two alleles of TIM in Drosophila species, variation of frequencies of this allele was shown across geographical location between Italy, Israel and Zimbabwe [68]. In our study, it remains puzzling that TIM showed population clustering within a village but not across villages. This may suggest genetic linkage between TIM and genes controlling mating incompatibility through cytological and temporal incompatibility of An. arabiensis population within a locality. It may be interesting to investigate how different alleles of TIM cluster across different geographical locations in the Kilombero valley to have a broader insight of the population structure of An. arabiensis within and between geographical localities. Further analysis based on larger set of markers or whole genome is required to unambiguously confirm of the effect of TIM on the observed clustering. However the existence of such fine-scale genetic structure as revealed by even the limited number of markers used here indicates there may be natural barriers to geneflow within An. arabiensis occurring over very small distances in the Kilombero Valley.
A few previous studies have investigated the population genetics of An. gambiae (s.l.) within the Kilombero Valley. One study similarly revealed the presence of strong structuring within An. arabiensis at the village-level in the Kilombero Valley, indicating this species exists in genetically distinct populations between villages situated only 40 km apart [63]. However in a recent study [72], An. arabiensis from the Kilombero Valley were predicted to exist within a single population including others from coastal Tanzania and the islands of Zanzibar. Further, a previous continental analysis of An. arabiensis population structure predicted relatively high levels of gene flow even between populations situated more than a 1000 km apart [73]. These contrasting findings may be partially due to limitations and discrepancies due to variation in the methods of analysis and selection of markers used.
Additional analyses of An. arabiensis population structure in our study using SNPs from mitochondrial genes has strengthened our arguments and those from previous work [63] indicate that An. arabiensis in the Kilombero valley may be divided into multiple subpopulations. While SNPs analysis from the clock genes showed possibility for two sub-populations of An. arabiensis that are not geographically related, analysis of mitochondrial SNPs has shown distinctive population structure within villages that are just 25 km apart (Fig. 4). These results in common strengthen the argument for sub-structure in An. arabiensis, which could be within or between localities in the Kilombero valley. This is an interesting phenomenon that requires further investigation using a broad range of markers to better understand the evolutionary processes in these vectors.
Future studies on the genetics of An. arabiensis behaviour should ideally involve a broader sampling strategy both with respect to the range and resolution of phenotypes selected, and the number of SNP markers used to increase the possibility of detecting genetic influences on their time and location of blood feeding. Further, it is possible that candidate clock genes may influence An. arabiensis feeding behaviour indirectly via epistatic interactions with other genes not considered here. To increase the power to detect both direct and epistatic genetic impacts on An. arabiensis, future studies using whole genome sequencing approaches are recommended. Additionally, transcriptomic approaches may also be useful given that they have been used successfully to detect associations between host choice behaviours and gene regulation in other insect vectors [74].
Conclusions
This study did not find any association between feeding behavioural phenotypes (early vs late and indoor vs outdoor feeding) in An. arabiensis and single nucleotide polymorphisms identified from eight candidate clock genes. However, there was evidence that the population contained two distinct genetic clusters that were associated with the Timeless gene, independently of feeding phenotype or geographical location. This finding was supported by analysis of eight mitochondrial genes, which showed that two lineages exist between two An. arabiensis populations just 25 km apart. It is highlighted that investigations of the genetic basis of the feeding behaviour in malaria vectors are still in their infancy, and will likely require much further development through use of high-resolution markers distributed across the entire genome, and/or the application of other methods including transcriptomic approaches to provide a strong test of genotype-phenotype associations. In studies where markers covering the whole genome have been applied, the high density and short LD in An. arabiensis observed implies that huge sample sizes could be needed to robustly test for such associations, which at present are not viable due to the high cost and time requirements. These limitations make a candidate gene approach more attractive in the short-term, however in future we recommend this could be improved through use of a larger set of genes, selected from across the entire genome of malaria vectors.
References
Gillies MT. The duration of the gonotrophic in Anopheles gambiae and Anopheles funestus, with a note on the efficiency of hand catching. East Afr Med J. 1953;30:129–35.
Gillies MT: The age-groups and the biting cycle in Anopheles gambiae. A preliminary investigation. Bull Entomol. Res, 1957;48:553–559 .
Mendis C, Jacobsen JL, Gamage-Mendis A, Bule E, Dgedge M, Thompson R, et al. Anopheles arabiensis and An. funestus are equally important vectors of malaria in Matola coastal suburb of Maputo, southern Mozambique. Med Vet Entomol. 2000;14:171–80.
Bryan JH, Petrarca V, Di Deco MA, Coluzzi M. Adult behaviour of members of the Anopheles gambiae complex in the Gambia with special reference to An. melas and its chromosomal variants. Parassitologia. 1987;29:221–49.
Smithuis FM, Kyaw MK, Phe UO, van der Broek I, Katterman N, Rogers C, et al. Entomological determinants of insecticide-treated bed net effectiveness in Western Myanmar. Malar J. 2013;12:364.
Oyewole IO, Awolola TS, Ibidapo CA, Oduola AO, Okwa OO, Obansa JA. Behaviour and population dynamics of the major anopheline vectors in a malaria endemic area in southern Nigeria. J Vector Borne Dis. 2007;44:56–64.
Maxwell CA, Wakibara J, Tho S, Curtis CF. Malaria-infective biting at different hours of the night. Med Vet Entomol. 1998;12:325–7.
White GB. Blood feeding habits of mosquitoes in the South Pare District of Tanzania ten years after cessation of a dieldrin residual spraying campaign. East Afr Med J. 1971;48:120–34.
Mahande A, Mosha F, Mahande J, Kweka E. Feeding and resting behaviour of malaria vector, Anopheles arabiensis with reference to zooprophylaxis. Malar J. 2007;6:100.
Githeko AK, Adungo NI, Karanja DM, Hawley WA, Vulule JM, Seroney IK, et al. Some observations on the biting behavior of Anopheles gambiae s.s., Anopheles arabiensis, and Anopheles funestus and their implications for malaria control. Exp Parasitol. 1996;82:306–15.
Braack LE, Coetzee M, Hunt RH, Biggs H, Cornel A, Gericke A. Biting pattern and host-seeking behavior of Anopheles arabiensis (Diptera: Culicidae) in northeastern South Africa. J Med Entomol. 1994;31:333–9.
Killeen GF, Chitnis N. Potential causes and consequences of behavioural resilience and resistance in malaria vector populations: a mathematical modelling analysis. Malar J. 2014;13:97.
Gordicho V, Vicente JL, Sousa CA, Caputo B, Pombi M, Dinis J, et al. First report of an exophilic Anopheles arabiensis population in Bissau City, Guinea-Bissau: recent introduction or sampling bias? Malar J. 2014;13:423.
Russell TL, Govella NJ, Azizi S, Drakeley CJ, Kachur SP, Killeen GF. Increased proportions of outdoor feeding among residual malaria vector populations following increased use of insecticide-treated nets in rural Tanzania. Malar J. 2011;10:80.
Fornadel CM, Norris LC, Glass GE, Norris DE. Analysis of Anopheles arabiensis blood feeding behavior in southern Zambia during the two years after introduction of insecticide-treated bed nets. Am J Trop Med Hyg. 2010;83:848–53.
Norris LC, Norris DE. Heterogeneity and changes in inequality of malaria risk after introduction of insecticide-treated bed nets in Macha, Zambia. Am J Trop Med Hyg. 2013;88:710–7.
Bayoh MN, Mathias DK, Odiere MR, Mutuku FM, Kamau L, Gimnig JE, et al. Anopheles gambiae: historical population decline associated with regional distribution of insecticide-treated bed nets in western Nyanza Provincem, Kenya. Malar J. 2010;9:62.
O’Meara WP, Mangeni JN, Steketee R, Greenwood B. Changes in the burden of malaria in sub-Saharan Africa. Lancet Infect Dis. 2010;10:545–55.
Derua YA, Alifrangis M, Hosea KM, Meyrowitsch DW, Magesa SM, Pedersen EM, et al. Change in composition of the Anopheles gambiae complex and its possible implications for the transmission of malaria and lymphatic filariasis in north-eastern Tanzania. Malar J. 2012;11:188.
Mwangangi JM, Mbogo CM, Orindi BO, Muturi EJ, Midega JT, Nzovu J, et al. Shifts in malaria vector species composition and transmission dynamics along the Kenyan coast over the past 20 years. Malar J. 2013;12:13.
Russell TL, Lwetoijera DW, Maliti D, Chipwaza B, Kihonda J, Charlwood JD, et al. Impact of promoting longer-lasting insecticide treatment of bed nets upon malaria transmission in a rural Tanzanian setting with pre-existing high coverage of untreated nets. Malar J. 2010;9:187.
Lounibos LP. Competitive displacement and reduction. J Am Mosq Control Assoc. 2007;23:276–82.
Charlwood JD, Vij R, Billingsley PF. Dry season refugia of malaria-transmitting mosquitoes in a dry savannah zone of east Africa. Am J Trop Med Hyg. 2000;62:726–32.
Okumu FO, Mbeyela E, Lingamba G, Moore J, Ntamatungiro AJ, Kavishe DR, et al. Comparative field evaluation of combinations of long-lasting insecticide treated nets and indoor residual spraying, relative to either method alone, for malaria prevention in an area where the main vector is Anopheles arabiensis. Parasit Vectors. 2013;6:46.
Matowo NS, Moore J, Mapua S, Madumla EP, Moshi IR, Kaindoa EW, et al. Using a new odour-baited device to explore options for luring and killing outdoor-biting malaria vectors: a report on design and field evaluation of the Mosquito Landing Box. Parasit Vectors. 2013;6:137.
Mayagaya VS, Nkwengulila G, Lyimo IN, Kihonda J, Mtambala H, Ngonyani H, et al. The impact of livestock on the abundance, resting behaviour and sporozoite rate of malaria vectors in southern Tanzania. Malar J. 2015;14:17.
Norris LC, Main BJ, Lee Y, Collier TC, Fofana A, Cornel AJ, et al. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc Natl Acad Sci U S A. 2015;112:815–20.
Ayala D, Ullastres A, Gonzalez J. Adaptation through chromosomal inversions in Anopheles. Front Genet. 2014;5:129.
Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Behavioural divergences between mosquitoes with different inversion karyotypes in polymorphic populations of the Anopheles gambiae complex. Nature. 1977;266:832–3.
Costantini C, Sagnon N, Ilboudo-Sanogo E, Coluzzi M, Boccolini D. Chromosomal and bionomic heterogeneities suggest incipient speciation in Anopheles funestus from Burkina Faso. Parassitologia. 1999;41:595–611.
Petrarca V, Beier JC. Intraspecific chromosomal polymorphism in the Anopheles gambiae complex as a factor affecting malaria transmission in the Kisumu area of Kenya. Am J Trop Med Hyg. 1992;46:229–37.
Mnzava AE, Mutinga MJ, Staak C. Host blood meals and chromosomal inversion polymorphism in Anopheles arabiensis in the Baringo District of Kenya. J Am Mosq Control Assoc. 1994;10:507–10.
Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95.
Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of clock in the Drosophila circadian oscillator. Neuron. 2003;37:249–61.
Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, et al. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–41.
Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol. 2011;62:335–64.
Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19:241–7.
Yuan Q, Metterville D, Briscoe AD, Reppert SM. Insect Cryptochromes: Gene Duplication and Loss Define Diverse Ways to Construct Insect Circadian Clocks. Mol Biol Evol. 2007;24:948–55.
Das S, Dimopoulos G. Molecular analysis of photic inhibition of blood-feeding in Anopheles gambiae. BMC Physiol. 2008;8:23.
Das S, Radtke A, Choi Y-J, Mendes AM, Valenzuela JG, Dimopoulos G. Transcriptomic and functional analysis of the Anopheles gambiae salivary gland in relation to blood feeding. BMC Genomics. 2010;11:566.
Rund SSC, Bonar NA, Champion MM, Ghazi JP, Houk CM, Leming MT, et al. Daily rhythms in antennal protein and olfactory sensitivity in the malaria mosquito Anopheles gambiae. Sci Rep. 2013;3:2494.
Rund SSC, Hou TY, Ward SM, Collins FH, Duffield GE. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc Natl Acad Sci U S A. 2011;108:E421–430.
Gross K, Alba S, Schellenberg J, Kessy F, Mayumana I, Obrist B. The combined effect of determinants on coverage of intermittent preventive treatment of malaria during pregnancy in the Kilombero Valley, Tanzania. Malar J. 2011;10:140.
Hetzel MW, Alba S, Fankhauser M, Mayumana I, Lengeler C, Obrist B, et al. Malaria risk and access to prevention and treatment in the paddies of the Kilombero Valley, Tanzania. Malar J. 2008;7:7.
Minja H, Schellenberg JA, Mukasa O, Nathan R, Abdulla S, Mponda H, et al. Introducing insecticide-treated nets in the Kilombero Valley, Tanzania: the relevance of local knowledge and practice for an information, education and communication (IEC) campaign. Trop Med Int Health. 2001;6:614–23.
Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–9.
Meireles-Filho ACA, Amoretty PR, Souza NA, Kyriacou CP, Peixoto AA. Rhythmic expression of the cycle gene in a hematophagous insect vector. BMC Mol Biol. 2006;7:38.
Rund SS, Gentile JE, Duffield GE. Extensive circadian and light regulation of the transcriptome in the malaria mosquito Anopheles gambiae. BMC Genomics. 2013;14:218.
Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–8.
Weyman PD, Pan Z, Feng Q, Gilchrist DG, Bostock RM. A circadian rhythm-regulated tomato gene is induced by Arachidonic acid and Phythophthora infestans infection. Plant Physiol. 2006;140:235–48.
Librado P, Rozas J. DnaSP v a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7.
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59.
Earl DA, Bridgett M. STRUCTURE Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;2:359–61.
Beard CB, Hamm DM, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 1993;2:103–24.
Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol. 2004;4:137–8.
Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc. 1922;85:87–94.
Dunn OJ. Multiple Comparisons Among Means. J Am Stat Assoc. 1936;56:52–64.
Peakall R, Smouse PE. GenAlEx 6. genetic analysis in Excel Population genetic software for teaching and researchan update. Bioinformatics. 2012;5:2537–9.
Marsden CD, Lee Y, Kreppel K, Weakley A, Cornel A, Ferguson HM, et al. Diversity, differentiation, and linkage disequilibrium: prospects for association mapping in the malaria vector Anopheles arabiensis. G3 (Bethesda). 2014;4:121–31.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Bioinformatics. 2013;00:1–3.
Lee Y, Seifert SN, Fornadel CM, Norris DE, Lanzaro GC. Single-nucleotide polymorphisms for high-throughput genotyping of Anopheles arabiensis in East and southern Africa. J Med Entomol. 2012;49:307–15.
Ng’habi KR, Knols BG, Lee Y, Ferguson HM, Lanzaro GC. Population genetic structure of Anopheles arabiensis and Anopheles gambiae in a malaria endemic region of southern Tanzania. Malar J. 2011;10:289.
Whitman DW, Ananthakrishnan TN: Phenotypic plasticity of insects: mechanisms and consequences, Science Publishers, 2009, 904.
Govella NJ, Okumu FO, Killeen GF. Insecticide-treated nets can reduce malaria transmission by mosquitoes which feed outdoors. Am J Trop Med Hyg. 2010;82:415–9.
Geissbuhler Y, Chaki P, Emidi B, Govella NJ, Shirima R, Mayagaya V, et al. Interdependence of domestic malaria prevention measures and mosquito-human interactions in urban Dar es Salaam, Tanzania. Malar J. 2007;6:126.
Yohannes M, Boelee E. Early biting rhythm in the Afro-tropical vector of malaria, Anopheles arabiensis, and challenges for its control in Ethiopia. Med Vet Entomol. 2012;26:103–5.
Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, et al. Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science. 2007;316:1895–8.
Rona LDP, Carvalho-Pinto CJ, Gentile C, Grisard EC, Peixoto AA. Assessing the molecular divergence between Anopheles cruzii populations from Brazil using the timeless gene: further evidence of a species complex. Malar J. 2009;8:60.
Silva-do-Nascimento TF, Pitaluga LDR, Peixoto AA, Lourenço-de-Oliveira R. Molecular divergence in the timeless and cpr genes among three sympatric cryptic species of the Anopheles triannulatus complex. Mem Inst Oswaldo Cruz. 2011;1:218–22.
Mathias D, Jacky L, Bradshaw WE, Holzapfel CM. Geographic and developmental variation in expression of the circadian rhythm gene, timeless, in the pitcher-plant mosquito, Wyeomyia smithii. J Insect Physiol. 2005;51:661–7.
Maliti D, Ranson H, Magesa S, Kisinza W, Mcha J, Haji K, et al. Islands and stepping-stones: comparative population structure of Anopheles gambiae sensu stricto and Anopheles arabiensis in Tanzania and implications for the spread of insecticide resistance. PLoS One. 2014;9:e110910.
Donnelly MJ, Licht MC, Lehmann T. Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles gambiae. Mol Biol Evol. 2001;18:1353–64.
McBride CS, Baier F, Omondi AB, Spitzer SA, Lutomiah J, Sang R, et al. Evolution of mosquito preference for humans linked to an odorant receptor. Nature. 2014;515:222–7.
Acknowledgements
We thank the European Union Commission for funding this research through the African Vector New Tools (AvecNet) project Work Package 7, sub-award number 201015556-2. Also we thank the NIH for funding this work through grant number R01AI085175. We thank Catelyn Neiman and Allison Weakley at the Vector Genetics Laboratory (UC Davis) for their assistance in training for DNA extraction, Qiaxel instrument handling, and iPLEX SNP genotyping preparations. We thank our field technicians Masoud Kilalangongono, Aniset Kihonda, Tokosa, Kasubiri and Tumpe Mwandyala. Finally, we acknowledge the contribution of a number of field volunteers and the community at Lupiro and Sagamaganga villages.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DM, GL, CM, HMF and YL conceived the study. DM conducted field collections, laboratory analyses and wrote the manuscript. YL designed multiplex SNP genotyping assay for both clock genes and mitochondrial genes. YY conducted mitochondrial SNP genotyping assay and scoring. TCC conducted genome sequence analysis. DM and YL conducted SNP genotyping and data analysis. HF, GL, BM, YY, TCC, YL, KK, NG, JCC, and CM reviewed the manuscripts. All authors read and approved the final version of the manuscript.
Additional files
Additional file 1: Table S4.
The SNP information including flanking sequences and primer sequences used for iPLEX SNP genotyping. (XLSX 13 kb)
Additional file 2: Figure S1.
Bayesian clustering analysis. The magnitude of ΔK as a function of K showing K = 2 as the most probable number of clusters when all 34 loci were included (A) and when only 4 Timeless loci were included in analysis (B). In (C) analysis was done on the remaining 30 with the exclusion of the Timeless loci. In D only the non-synomymous SNPs were analys ed. Assumption of 8 populations was made apriori. The 8 populations were assumed according the 8 feeding phenotypes, which included 4 phenotypes from Lupiro and 4 from Sagamaganga (PDF 1039 kb)
Additional file 3: Table S2.
Nucleotide sequences on the 8 genes from mitochondria that distinguish two lineages (MINEPA and SAGAMAGANGA) in Tanzania. (DOCX 11 kb)
Additional file 4: Table S3.
Sample size and Chi-square test results between the feeding phenotypes and the sampling sites of Lupiro and Sagamaganga. (DOCX 11 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Maliti, D.V., Marsden, C.D., Main, B.J. et al. Investigating associations between biting time in the malaria vector Anopheles arabiensis Patton and single nucleotide polymorphisms in circadian clock genes: support for sub-structure among An. arabiensis in the Kilombero valley of Tanzania. Parasites Vectors 9, 109 (2016). https://doi.org/10.1186/s13071-016-1394-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1394-8