Abstract
Background
African animal trypanosomiasis (AAT) is considered to be one of the greatest constraints to livestock production and livestock-crop integration in most African countries. South-eastern Uganda has suffered for more than two decades from outbreaks of zoonotic Human African Trypanosomiasis (HAT), adding to the burden faced by communities from AAT. There is insufficient AAT and HAT data available (in the animal reservoir) to guide and prioritize AAT control programs that has been generated using contemporary, sensitive and specific molecular techniques. This study was undertaken to evaluate the burden that AAT presents to the small-scale cattle production systems in south-eastern Uganda.
Methods
Randomised cluster sampling was used to select 14% (57/401) of all cattle containing villages across Tororo District. Blood samples were taken from all cattle in the selected villages between September-December 2011; preserved on FTA cards and analysed for different trypanosomes using a suite of molecular techniques. Generalized estimating equation and Rogen-Gladen estimator models were used to calculate apparent and true prevalences of different trypanosomes while intra cluster correlations were estimated using a 1-way mixed effect analysis of variance (ANOVA) in R statistical software version 3.0.2.
Results
The prevalence of all trypanosome species in cattle was 15.3% (95% CI; 12.2-19.1) while herd level trypanosome species prevalence varied greatly between 0-43%. Trypanosoma vivax (17.4%, 95% CI; 10.6-16.8) and Trypanosoma brucei rhodesiense (0.03%) were respectively, the most, and least prevalent trypanosome species identified.
Conclusions
The prevalence of bovine trypanosomes in this study indicates that AAT remains a significant constraint to livestock health and livestock production. There is need to implement tsetse and trypanosomiasis control efforts across Tororo District by employing effective, cheap and sustainable tsetse and trypanosomiasis control methods that could be integrated in the control of other endemic vector borne diseases like tick-borne diseases.
Similar content being viewed by others
Background
African Animal Trypanosomiasis (AAT) is one of the most important vector-borne diseases of livestock in East Africa and common throughout the tsetse belts of Africa [1],[2]. Trypanosoma congolense, Trypanosoma vivax and Trypanosoma brucei subspecies brucei are the most important causes of AAT mainly transmitted by tsetse flies (Glossina ssp.) [3]. AAT is usually chronic; characterized by loss of condition, progressive anaemia and often terminates in death if untreated [4]. T. vivax can also be transmitted mechanically by broad spectra of haematophagous insects and as a result, AAT caused by T. vivax has been recorded outside of the tsetse belts [5]. T. brucei rhodesiense and T.b. brucei are less pathogenic in cattle than T.congolense and T. vivax[5]. The persistent and long-term presence of T. brucei rhodesiense in cattle as a reservoir of human infection is of major public health importance [6] with spillover from domestic livestock causing human T.b. rhodesiense African trypanosomiasis (HAT), also known as sleeping sickness [7]-[9]. Domestic animals of epidemiological importance, are, notably cattle, which act as reservoirs of the human infective trypanosomes [10]-[12].
In Sub-Saharan Africa and south-eastern Uganda in particular, vector-borne diseases notably AAT constrain livestock production and compound poverty levels contributing to 34% of all livestock keepers to subsist on less than 1.24 USD per day [13]-[15]. Poor livestock health as a result of AAT denies farmers the opportunity to use draft power and manure as the gateway to crop-livestock enterprise integration thereby extending this problem of poverty and hunger in the tsetse-infested areas [1],[2]. Hunger continues to affect more than one third of the populations in this region [16],[17].
There is little contemporary data available on AAT burden generated from the sensitive and specific molecular techniques that are widely available. Such data is essential for guiding and prioritising tsetse control and the integration of such control efforts with those of other endemic vector-borne diseases such as tick-borne diseases (TBDs). The present study represents the first large scale attempt to generate reliable prevalence data from which to evaluate the extent to which the African trypanosomiasis presents a problem in small scale cattle production systems in south-eastern Uganda. A cross sectional survey was undertaken to determine both the prevalence and spatial distribution of African trypanosomes across Tororo District.
Methods
Study area
The survey was carried out in Tororo during the period September to December 2011. Tororo District is bordered by the districts of Mbale to the north, Manafwa to the north-east, Busia to the south, Bugiri to the south-west, Butaleja to the north-west and the Republic of Kenya to the east. The location, farming system, climate and vegetation of the study area have been previously described [18]-[20]. Cattle are the main tsetse hosts in Tororo district [11],[12] contributing up to 54% of all tsetse blood meals with the rest taken from pigs and monitor lizards (Varanus niloticus) [12]. Glossina fuscipes fuscipes and G. pallidipes are the main tsetse fly species in the district [12]. At the time of the study, the district had estimated cattle population of 37,345 in 401 villages (average = 93 cattle/village) [21]. There was no evidence of any mass treatment of cattle against AAT in Tororo District between 2010–2011.
Study design and sampling methods
A cross sectional study was carried out involving taking of blood samples from all cattle in 57 of the 401 cattle containing villages of Tororo District. The cluster sampling methodology [22],[23] implemented in C Survey version 2.0 [24] was used to determine the minimum number of clusters needed. Cluster sampling allows variation in the number of clusters and cattle that are sampled (e.g., some cattle in all clusters, all cattle in all clusters; all cattle in some clusters) to achieve the specified parameters. Owing to the large size of this survey, village selection was done by simple random sampling to reduce sampling errors that often result from inherent variability between different samples drawn from a large population (sampling frame). The sampling frame; comprising a complete list of all villages and their geo-referenced positions in Tororo District was obtained from the Coordinating Office for Control of Trypanosomiasis in Uganda (COCTU) and verified for completeness at Tororo District lands and planning offices.
Sample size was determined assuming a mean cattle population of 93 cattle per village [21], anticipated prevalence of AAT of 30% based on the experience of the investigators, and some published general trypanosome studies [25], the precision of the sample estimate (one half-length of the 95% confidence interval) of 5 percentage points and an intracluster correlation coefficient (ICC) of 0.15. The ICC estimate was based on reported rates of homogeneity (rho) for trypanosomiasis prevalence, noting high variability [26],[27]. The number of cattle expected to be sampled per cluster was taken as the mean of the number of cattle per village in Tororo District (93). In total, fifty seven (57) clusters (villages) were needed to achieve the set level of precision for trypanosome prevalence estimation. The number of clusters selected fulfil the sampling assumption that the cluster means are normally distributed [28] indicating that a minimum of 30 clusters could have been used.
Infectious diseases (both vector-borne and non-vector-borne) display heterogeneity within a population; that is, they tend to cluster into epidemiological units; a group of animals that is of epidemiological significance in terms of the transmission and maintenance of infection, and therefore of disease control. For this reason, the sampling units/clusters for this study are villages; the epidemiological units for trypanosomiasis. A livestock containing village in Tororo District is here defined as a cluster because, in communal husbandry obtaining this is the epidemiological unit.
Cattle blood sample collection
About 125 μl of blood was collected from the middle ear vein and applied onto a designated sample area of the classic Flinders Technology Associates (FTA®) cards (Whatman Bioscience, Cambridge, UK), avoiding cross contamination [29],[30]. Blood samples were then allowed to air-dry, labelled with village name, parish, sub county, county and date of collection. They were packed in foil pouches with a silica gel desiccant (Sigma Aldrich, Co., Life sciences, USA) prior to shipping to the University of Edinburgh, UK for analysis.
DNA extraction
DNA was extracted and eluted in Chelex®100 resin (Sigma Aldrich, Co., Life sciences, USA) from five 3 mm FTA sample discs according to a previously described protocol [30],[31]. Eluted DNA samples were kept at −20°C for long term PCR analyses or 4°C if they were to be analysed within a few days after extraction.
Trypanosome DNA detection
Eluted DNA samples were screened for different trypanosome species using a single pair of primers (CR and BR) previously designed to amplify internal transcribed spacer (ITS1) of trypanosome ribosomal deoxyribonucleic acid (rDNA) and thermo cycling conditions as previously described [32]. The ITS1- PCR was carried out in a 25 μl reaction volume; 20 μl of which was the PCR master mix and either 5 μl of the test sample or negative control eluate or positive control DNA. The master mix was made of 10x-reaction buffer (670 mM Tris–HCl pH 8.8, 166 μM (NH4)2SO4, 4.5% Triton X-100, 2 mg/ml gelatin) (Fisher Biotech), 1.0 mM MgCl2, 200 μM of each dNTP, 5 μM each of the CF and BR primers, 0.5U of Taq DNA polymerase (Fisher Biotech) and 15.2 μl RNase-free molecular grade water.
To determine which samples were infected with either T. brucei or T. b. rhodesiense, multiplex PCR [33] was carried out on each of the samples from which a 450 bp fragment was detected on ITS1-PCR. Multiplex PCR was carried out in 25 μl reactions using primers and conditions as previously described [33]
To determine the commonest T.congolense genotype circulating in Tororo District, all samples from which a ≥600 bp fragment was amplified on ITS1-PCR were initially tested for T.congolense savannah using a single pair of primers (TCS1 & TCS2) and thermo cycling conditions as previously described [34]. All samples that were positive for T.congolense DNA on ITS1-PCR were positive for T.congolense savannah. For this reason, no more T.congolense genotype-specific (kilifi, tsavo, forest) PCRs were performed although a few co-infections with different T.congolense genotypes could have been possible. The PCR was carried out in a 25 μl reaction volume; 20 μl of which was the PCR master mix and either 5 μl of the test sample or negative control eluate or positive control DNA. The master mix was made of 10x-reaction buffer (670 mM Tris–HCl pH 8.8, 166 μM(NH4)2SO4, 4.5% Triton X-100, 2 mg/ml gelatin) (Fisher Biotech),, 4.5% Triton X-100, 2 mg/ml gelatin) (Fisher Biotech), 0.75 mM MgCl2, 200 μM of each dNTP, 12.5 μM each of the TCS1 & TCS2 primers, 1U of Taq DNA polymerase (Fisher Biotech) and 13.05 μl of RNase-free water.
PCR products for the three sets of PCRs were separated in 1.5% agarose (Bio Tolls Inc. Japan), stained in GelRed™ (Biotium, Inc., USA) and visualised on an ultraviolet transilluminator for fragment size determination
Statistical analysis
Apparent prevalences and their confidence intervals were estimated using the generalized estimating equation models to adjust for correlations within communities. True prevalences were calculated using the Rogen-Gladen estimator. Intra cluster correlations were estimated using a 1-way mixed effect ANOVA model. All statistical analyses were performed using the R statistical software version 3.0.2. ArcMap v10.3 was used to map prevalence estimates in different villages.
Ethical clearance
This study was reviewed by the Makerere University College of Veterinary Medicine Animal Resources and Biosecurity ethical review board for compliance to Animal use and Care standards. It was then forwarded to the Uganda National Council for Science and Technology and approved under approval number HS1336.
Results
Demographic characteristics of the study population
Six thousand fifty four blood samples were taken from all cattle in 57 villages in Tororo District. The mean number of cattle per village was 106 (4–232). The mean number of cattle per household was 4. The demographic characteristics of the study population are summarized in Table 1. Almost all the animals belonged to the Boran × short horn Zebu cross-breed. Approximately half of the population were female.
Bovine trypanosome species prevalence
Out of 6,054 cattle sampled, 850 were infected with a single trypanosome species and a further 78 animals had mixed trypanosome infections (Table 2). The most common co-infections observed were T. vivax and T. b. brucei (37 animals) and T. vivax and T. congolense (34 animals). The overall prevalence of different trypanosome species was 15.3% (95% CI; 12.2-19.1%). T.brucei s.l. constituted the smallest proportion of trypanosomes in cattle in Tororo District.
Herd level prevalence of different bovine trypanosome species
There was a very large variation in the prevalence of different trypanosome species between different villages (clusters) with some villages recording 0 infection rates while others recording very high infection rates of up to 43% (Table 3). As a result, the rate of homogeneity (roh)/ intra-cluster correlation coefficient (ICC) for any trypanosome infection was estimated at 0.11. The most common trypanosome species was T. vivax with an apparent prevalence of 13.4 (10.6-16.8) occurring in 52 out of 57 communities respectively. T. c. savannah and T.brucei s.l. were detected in 53% and 35% of all villages sampled. T.b. rhodesiense was detected only in 2 of the 57 (3.5%) villages sampled.
Prevalence of T.brucei s.l
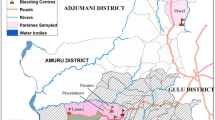
Only 69 blood samples were positive for T.brucei s.l. (Figure 1). Further characterisation by multiplex PCR identified 2 of the 69 samples as positive for the human infective T. b. rhodesiense. The two T. b. rhodesiense positive samples were from the villages of Chawolo-Sironga B and Kadanya; one positive sample from each of the two villages
Prevalence of T.brucei s.l. in cattle in 57 villages of Tororo District, Uganda. T.brucei s.l. prevalence was categorized into five classes for which symbols differ in size and colour. County boundaries were included for ease of assessment of the location of sample sites within the district. The estimated number of cases per 100 animals is presented within the symbols. Only names of villages with the highest prevalence estimates are added.
Spatial distribution of bovine trypanosomes
The lowest prevalences were registered along the Kenyan Border while the highest prevalences were recorded in villages in the northern and the western parts of the district bordering the districts of Manafwa and Mbale (Figure 2).
Spatial distribution of bovine trypanosome species and land cover. The overall prevalence of different trypanosome species in each village was categorized into five classes for which symbols differ in size and colour. County boundaries were included for ease of assessment of the location of sample sites within the district. The estimated number of cattle infected with different trypanosome species per 100 animals are presented within the symbols. Only name labels of villages with the highest prevalence estimates were added to avoid overcrowding the map. A background layer of land cover classes (GLC2000) was added to assess the likely effect of land use on trypanosome prevalence.
Discussion
To determine the burden and spatial distribution of African trypanosomes in crop-livestock production systems in Tororo District, south-eastern Uganda, cattle blood samples were taken and tested for different trypanosomes between September and December 2011. Six thousand and fifty four cattle blood samples from 57 villages were analysed for different trypanosomes. The overall prevalence for different trypanosomes in Tororo District of 15.3% (95% CI; 12.2-19.1%) was comparable to those previously reported in this region [12],[35]. Individual species prevalences were higher than those reported in previous studies [12],[35]. The most pathogenic species for cattle, T. vivax and T. congolense were recorded at 13.4% (95% CI; 10.6-16.8) and 2.1% (95% CI; 1.4-3.1) respectively. T. brucei s.l was observed at a prevalence of 1.1% (95% CI; 0.7-1.8) with a low frequency [36] with 2 of 69 positive samples being positive for SRA gene, indicative of human infectivity [10].
The prevalence of different trypanosome species greatly varied between different villages ranging from 0-43% (Table 3). Villages with medium to high AAT prevalence were clustered along the northern and western borders of Tororo District along the Kenyan border (Figure 2). These areas are mainly covered by forest/savannah vegetation and croplands interspersed with cattle which make the ideal conditions for tsetse infestation. Trypanosome prevalence and land use showed a positive spatial association, which might explain the high prevalences found in the north and western boarders of Kenya (Figure 2). This could be as a result of the differences in tsetse apparent density and veterinary care between villages [37].
T. vivax was detected in 91% of all sampled villages. Glossina fuscipes fuscipes has been reported to be the commonest tsetse fly species in eastern Uganda [38]-[40] and was the commonest tsetse species caught in a recent survey at 161 locations in Tororo District [37]. G. f. fuscipes is a better vector for T.vivax than G. pallidipes[37],[40], which is scarce despite recent re-invasion in this part of Uganda [35]. This aside, mechanical transmission may explain why T. vivax was detected in most sampled villages. T. vivax undergoes a short life cycle in the proboscis of tsetse [42]-[44] and a fast build-up of parasitaemia in mammalian hosts [45] which may contribute to T.vivax detection in cattle.
Prevalences of T. congolense, T.b. brucei and T. b. rhodesiense in Tororo District were lower than observed for T. vivax. However, T. c. savannah was detected in slightly above half (53%) of all the herds (villages) screened. The wider distribution of T. c. savannah in Tororo District herds may be as a result of re-invasion of G.pallidipes in Tororo, associated with risk of transmission of T. c. savannah[38].
T. congolense and the haemorrhagic strain of T.vivax are the most pathogenic trypanosome species of cattle in East Africa causing an acute and fatal disease compared to a more chronic form of AAT caused by other bovine trypanosome species and strains [34],[46]. That T. congolense infections progress to a fatal disease rapidly, demanding treatment, may explain why T. congolense was detected in low prevalences in apparently healthy animals screened in the 57 villages of Tororo [47]. The very high prevalence and wide distribution of T. congolense and T.vivax in Tororo District indicates that these highly pathogenic trypanosome species continue to constrain livestock production calling for sustainable trypanosomiasis control measures.
T. b brucei and T. b. rhodesiense were detected in low individual species and herd prevalences. The very low prevalence (0.03%) of T. b. rhodesiense in cattle indicates that there still remains a low risk of transmission of T. b. rhodesiense from cattle to humans. This is especially so in the two villages of Chawolo-Sironga B and Kadanya where two cattle blood samples were found positive for T. b. rhodesiense, one sample from each village. Given that ITS1-PCR has slightly lower sensitivity (95%) to T.brucei s.l. than the species specific PCR (~100%) T.brucei s.l. may have been under detected by ITS1-PCR [48]-[50].
The last outbreak of sleeping sickness in Tororo District was in 2000/2001 [50]. Since then, there have not been reports of the human disease in the district. That T. b. rhodesiense was detected only in 2 out of 6,054 cattle sampled shows that the cattle reservoir for this infection in Tororo District is persistent and is of concern. The presence of human infective T. b. rhodesiense animal carriers within isolated villages in Tororo District indicates that humans in these isolated villages remain at risk of acquiring infection given the abundance of tsetse flies [51].
Cattle are grazed communally in large herds (average of 106) in Tororo but at night are kept in smaller groups (average of 4) in night-shades or tethered around homesteads [19],[20],[37]. The number of animals per household is low typical of livestock holding in south-eastern Uganda. Boran and African short horn Zebu (Nkedi) hybrids and Nkedi are the major cattle breeds kept in Tororo District [19],[20],[52]. The male to female cattle ratio is high (0.8) in Tororo, since farmers need to retain bulls (whole or neutered) for 3 years or more to provide draught power in these mixed crop-livestock systems [19],[53]. Cattle over three years of age have been associated with a higher risk of infection and spread of human infective T.b rhodesiense[54]. Production systems that retain a very high proportion of cattle above 3 years of age pose a risk for zoonotic T.b. rhodesiense HAT transmission. Improving livestock health by controlling tsetse and trypanosomiasis will reduce HAT incidence, enhance livestock production and livestock-crop integration thereby reducing poverty and hunger [17],[55]-[57]
The individual and herd prevalences reported in this study show that African trypanosomiasis remains a constraint to livestock production and livestock-crop integration in Tororo District despite farmer and local government-led tsetse and trypanosomiasis control efforts [36]. Lack of sustainability caused by insufficient follow-up, civil unrest and inadequate financing have resulted in failure of most tsetse control programs in Uganda [58]. Cattle as persistent reservoirs of human HAT should be treated to remove risk to poor communities in affected districts [6]. To prevent reinfection by tsetse control, the control methods used ought to be effective and sustainable. This requires that such methods are tailored to the limited budgets of poor rural livestock keepers and are effective against multiple endemic livestock diseases [59]. Control methods based on the use of restricted application of insecticides (RAP) on predilection sites for tsetse and/or ticks or spraying larger animals could serve to reduce the cost of RAP and offer added value of targeting ticks and biting flies making it easily adoptable for routine tsetse control [60]-[62].
Conclusion
African animal trypanosomiasis continues to be one of the main constraints to livestock production in Uganda. The current study indicates that the prevalence of different trypanosome species in Tororo District is still high despite government and farmer-led tsetse and trypanosomiasis control efforts. There is need to further intensify tsetse and trypanosomiasis control efforts in the district preferably by employing effective and sustainable tsetse and trypanosomiasis control methods. Such methods would need to be compatible to inelastic budgets of resource poor livestock keepers and be effective against most important endemic vector-borne diseases namely; tsetse and tick-borne diseases.
References
Murray M, Gray AR: The current situation on animal trypanosomiasis in Africa. Prev Vet Med. 1984, 2: 23-30. 10.1016/0167-5877(84)90045-X.
Swallow B: Impacts of African animal trypanosomosis on migration, livestock and crop production.Nairobi, ILRI 1998:1-19.,
Clarkson MJ: Trypanosomes. Vet Parasitol. 1976, 2 (1): 9-29. 10.1016/0304-4017(76)90050-9.
Magona JW, Mayende JS, Olaho-Mukani W, Coleman PG, Jonsson NN, Welburn SC, Eisler MC: A comparative study on the clinical, parasitological and molecular diagnosis of bovine trypanosomosis in Uganda. Onderstepoort J Vet Res. 2003, 70 (3): 213-218.
Shaw AP: Assessing the economics of animal trypanosomosis in Africa-history and current perspectives. Onderstepoort J Vet Res. 2009, 76 (1): 27-32. 10.4102/ojvr.v76i1.57.
Fevre EM, Coleman PG, Odiit MD, Magona J, Welburn SC MEJ: Woolhouse The origins of a new sleeping sickness outbreak (caused by Trypanosoma brucei infection) in eastern Uganda. Lancet. 2001, 358: 625-628. 10.1016/S0140-6736(01)05778-6.
Wardrop N, Fevre EM, Atkinson P, Welburn SC: The dispersal ecology of Rhodesian sleeping sickness following its introduction to a new area. PLoS Negl Trop Dis. 2013, 7 (10): e2485-10.1371/journal.pntd.0002485.
Selby R, Bardosh K, Waiswa C, Welburn SC: Cattle movements and trypanosomes: Restocking efforts and the spread of Rhodesian sleeping sickness in post-conflict Uganda. Parasit Vectors. 2013, 6: 281-10.1186/1756-3305-6-281.
Desquesnes M, Dia ML: Mechanical transmission of Trypanosoma vivax in cattle by the African tabanid Atylotus fuscipes. Vet Parasitol. 2004, 119 (1): 9-19. 10.1016/j.vetpar.2003.10.015.
Welburn SC, Picozzi K, Fevre EM, Coleman PG, Odiit M, Carrington M, Maudlin I: Identification of human infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet. 2001, 358: 2017-2019. 10.1016/S0140-6736(01)07096-9.
Picozzi K, Fevre EM, Odiit M, Carrington M, Eisler MC, Maudlin I, Welburn SC: Sleeping sickness in Uganda: a thin line between two fatal diseases. BMJ. 2005, 331 (7527): 1238-1241. 10.1136/bmj.331.7527.1238.
Waiswa C, Olaho-Mukani W, Katunguka-Rwakishaya E: Domestic animals as reservoirs for sleeping sickness in three endemic foci in south-eastern Uganda. Ann Trop Med Parasitol. 2003, 97 (2): 149-155. 10.1179/000349803235001688.
Minjauw B, McLeod A: Tick-borne diseases and poverty. The impact of ticks and tick- borne diseases on the livelihood of small-scale and marginal livestock owners in India and eastern and southern Africa. Research report, DFID Animal Health Programme Centre for Tropical Veterinary Medicine, University of Edinburgh, UK
Perry B, Sones K: Poverty reduction through animal health. Science. 2007, 315 (5810): 333-334. 10.1126/science.1138614.
Kabayo JP: Aiming to eliminate tsetse from Africa. Trends Parasitol. 2002, 18 (11): 473-475. 10.1016/S1471-4922(02)02371-1.
Hotez PJ: The neglected tropical diseases and their devastating health and economic impact on the member nations of the Organisation of the Islamic Conference. PLoS Negl Trop Dis. 2009, 3 (10): e539-10.1371/journal.pntd.0000539.
Hursey BSJ: Slingernbergh: The tsetse fly and its effects on agriculture in sub-Saharan Africa. World Anim Rev. 1995, 84–85: 67-73.
Wardrop NA, Fèvre EM, Atkinson PM, Kakembo A, Welburn SC: An exploratory GIS-based method to identify and characterise landscapes with an elevated epidemiological risk of Rhodesian human African trypanosomiasis. BMC Infect Dis. 2012, 12: 316-10.1186/1471-2334-12-316.
Muhanguzi D, Picozzi K, Hatendorf J, Thrusfield M, Welburn SC, Kabasa JD, Waiswa C: Prevalence and spatial distribution of Theileria parva in cattle under crop-livestock farming systems in Tororo District, Eastern Uganda. Parasit Vectors. 2014, 7: 91-10.1186/1756-3305-7-91.
Muhanguzi D, Picozzi K, Hatendorf J, Thrusfield M, Welburn SC, Kabasa JD, Waiswa C: Collateral benefits of restricted insecticide application for control of African trypanosomiasis on Theileria parva in cattle: a randomized controlled trial. Parasit Vectors. 2014, 7 (1): 432-10.1186/1756-3305-7-432.
Anonymous: The National Livestock Census: A summary report of the national livestock census. Kampala: Uganda Bureau of Statistics: Ministry of Agriculture Animal Industry and Fisheries Kampala, 2009:41.
Bennett S, Woods T, Liyanage WM, Smith DL: A simplified general method for cluster-sample surveys of health in developing countries. World Health Stat Q. 1991, 44 (3): 98-106.
Thrusfield M: Veterinary Epidemiology. 2007, Blackwell science, Oxford, 3
Farid MN, Frerichs RR: CSurvey, version 2.0. Department of Epidemiology, University of California, Los Angels (UCLA), Los Angeles. 2007.
Bohning D, Greiner M: Prevalence estimation under heterogeneity in the example of bovine trypanosomosis in Uganda. Prev Vet Med. 1998, 36 (1): 11-23. 10.1016/S0167-5877(98)00076-2.
Orjuela J, Navarrete M, Betancourst A, Roqueme L, Cortez E, Morrison RB: Salud y productividad en bovines de la costa norte en Colombia. 2. Hallazgos, serologicos, bacteriologicos y parasitologicos. World Anim Rev. 1991, 69: 7-14.
Dhalwa J: Animal Health and Production Constraints in Traditional Farming Systems in Mbarara District, Uganda. University of Reading, UK; 1995.
Cochran WG: Sampling Techniques. 1977, Wiley, New York, 3
Picozzi K, Tilley A, Fèvre E, Coleman P, Magona J, Odiit M, Eisler M, Welburn S: The diagnosis of trypanosome infections: applications of novel technology for reducing disease risk. Afr J Biotechnol. 2002, 1 (2): 39-45.
Ahmed HA, MacLeod ET, Hide G, Welburn SC, Picozzi K: The best practice for preparation of samples from FTA(R)cards for diagnosis of blood borne infections using African trypanosomes as a model system. Parasit Vectors. 2011, 4: 68-10.1186/1756-3305-4-68.
Becker S, Franco JR, Simarro PP, Stich A, Abel PM, Steverding D: Real-time PCR for detection of Trypanosoma brucei in human blood samples. Diagn Microbiol Infect Dis. 2004, 50 (3): 193-199. 10.1016/j.diagmicrobio.2004.07.001.
Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RC, Davila AM: The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005, 95 (3): 186-192. 10.1007/s00436-004-1267-5.
Picozzi K, Carrington M, Welburn SC: A multiplex PCR that discriminates between Trypanosoma brucei brucei and zoonotic T. b. rhodesiense. Exp Parasitol. 2008, 118 (1): 41-46. 10.1016/j.exppara.2007.05.014.
Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC: Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992, 22 (7): 909-918. 10.1016/0020-7519(92)90047-O.
Magona JW, Walubengo J, Odiit M, Okedi LA, Abila P, Katabazi BK, Gidudu AM, Olaho-Mukani W: Implications of the re-invasion of Southeast Uganda by Glossina pallidipes on the epidemiology of bovine trypanosomosis. Vet Parasitol. 2005, 128 (1–2): 1-9. 10.1016/j.vetpar.2004.10.020.
Coleman PG, Welburn SC: Are fitness costs associated with resistance to human serum in Trypanosoma brucei rhodesiense?. Trends Parasitol. 2004, 20: 311-315. 10.1016/j.pt.2004.04.015.
Muhanguzi D, Picozzi K, Hatendorf J, Thrusfield M, Welburn SC, Kabasa JD, Waiswa C: Improvements on restricted insecticide application protocol for control of human and animal african trypanosomiasis in eastern Uganda. PLoS Negl Trop Dis. 2014, 8 (10): e3284-10.1371/journal.pntd.0003284.
Moloo SK, Kutuza SB, Boreham PF: Studies on Glossina pallidipes. G. fuscipes fuscipes and G. brevipalpis in terms of the epidemiology and epizootiology of trypanosomiases in south-eastern Uganda. Ann Trop Med Parasitol. 1980, 74 (2): 219-237.
Okoth JO: Observation on the composition of Glossina population at Lugala, south Busoga district, Uganda. East Afr Med J. 1980, 57: 332-336.
Okoth JO: Further observations on the composition of glossina population at Lugala, South Busoga, Uganda. East Afr Med J. 1982, 59: 582-584.
Moloo S: Interacting factors in the epidemiology of trypanosomiasis in an endemic/enzootic region of Uganda and its contiguous area of Kenya. Insect Sc Appl. 1980, 1 (01): 117-121.
Gardiner PR, Wilson AJ: Trypanosoma (Duttonefla) vivax. Parasitol Today. 1987, 3 (2): 49-52. 10.1016/0169-4758(87)90213-4.
Jones TW, Davila AM:Trypanosoma vivax-out of Africa. Trends Parasitol. 2001, 17 (2): 99-101. 10.1016/S1471-4922(00)01777-3.
Jefferies D, Helfrich MP, Molyneux DH: Cibarial infections of Trypanosoma vivax and T. congolense in Glossina. Parasitol Res. 1987, 73 (4): 289-292. 10.1007/BF00531079.
Osório AL, Madruga CR, Desquesnes M, Soares CO, Ribeiro LR, Costa SC: Trypanosoma (Duttonella) vivax: its biology, epidemiology, pathogenesis, and introduction in the New World - a review. Mem Inst Oswaldo Cruz. 2008, 103: 1-13. 10.1590/S0074-02762008000100001.
Magona JW, Walubengo J, Odimin JT: Acute haemorrhagic syndrome of bovine trypanosomosis in Uganda. Acta Trop. 2008, 107 (2): 186-191. 10.1016/j.actatropica.2008.05.019.
Leak SGA, Rowlands GJ: The dynamics of trypanosome infections in natural populations of tsetse (Diptera: Glossinidae) studied using wing-fray and ovarian ageing techniques. Bull Entomol Res. 1997, 87 (03): 273-282. 10.1017/S0007485300037226.
Cox A, Tosas O, Tilley A, Picozzi K, Coleman P, Hide G, Welburn S: Constraints to estimating the prevalence of trypanosome infections in East African zebu cattle. Parasit Vectors. 2010, 3: 82-10.1186/1756-3305-3-82.
Moser DR, Kirchhoff LV, Donelson JE: Detection of Trypanosoma cruzi by DNA amplification using the polymerase chain reaction. J Clin Microbiol. 1989, 27 (7): 1477-1482.
Ahmed HA, Picozzi K, Welburn SC, MacLeod ET: A comparative evaluation of PCR- based methods for species- specific determination of African animal trypanosomes in Ugandan cattle. Parasit Vectors. 2013, 6 (1): 316-10.1186/1756-3305-6-316.
Odiit M, Bessell PR, Fevre EM, Robinson T, Kinoti J, Coleman PG, Welburn SC, McDermott J, Woolhouse ME: Using remote sensing and geographic information systems to identify villages at high risk for rhodesiense sleeping sickness in Uganda. Trans R Soc Trop Med Hyg. 2006, 100 (4): 354-362. 10.1016/j.trstmh.2005.04.022.
Magona JW, Walubengo J, Odimim JJ: Differences in susceptibility to trypanosome infection between Nkedi Zebu and Ankole cattle, under field conditions in Uganda. Ann Trop Med Parasitol. 2004, 98 (8): 785-792. 10.1179/000349804225021532.
Ocaido M, Otim CP, Okuna NM, Erume J, Ssekitto C, Wafula RZO, Kakaire D, Walubengo J, Monrad J: Socio-economic and livestock disease survey of agro-pastoral communities in Serere County, Soroti District, Uganda. Livest Res Rural Dev. 2005, 17: 93-
von Wissmann B, Fyfe J, Picozzi K, Hamill L, Waiswa C, Welburn SC: Quantifying the association between bovine and human trypanosomiasis in newly affected sleeping sickness areas of Uganda. PLoS Negl Trop Dis. 2014, 8 (6): e2931-10.1371/journal.pntd.0002931.
Murray M, Gray A: The current situation on animal trypanosomiasis in Africa. Prev Vet Med. 1984, 2 (1): 23-30. 10.1016/0167-5877(84)90045-X.
Swallow BM: Impacts of Trypanosomiasis on African Agriculture. 1999, FAO, Rome
Machila N, Emongor R, Shaw AP, Welburn SC, McDermott J, Maudlin I, Eisler MC: A community education intervention to improve bovine trypanosomiasis knowledge and appropriate use of trypanocidal drugs on smallholder farms in Kenya. Agr Syst. 2007, 94 (2): 261-272. 10.1016/j.agsy.2006.09.004.
Allsopp R: Options for vector control against trypanosomiasis in Africa. Trends Parasitol. 2001, 17 (1): 15-19. 10.1016/S1471-4922(00)01828-6.
Okello A, Welburn S, Kabasa JD, Waiswa C, Rannaleet A, Mitchell M, Semakula L: Stamp Out Sleeping Sickness (SOS): An Innovative One Health Approach to Neglected Zoonotic Disease in Uganda. Ecohealth. 2011, 7: S72–S72-
Hargrove JW, Ouifki R, Kajunguri D, Vale GA, Torr SJ: Modeling the control of trypanosomiasis using trypanocides or insecticide-treated livestock. PLoS Negl Trop Dis. 2012, 6 (5): e1615-10.1371/journal.pntd.0001615.
Torr SJ, Maudlin I, Vale GA: Less is more: restricted application of insecticide to cattle to improve the cost and efficacy of tsetse control. Med Vet Entomol. 2007, 21 (1): 53-64. 10.1111/j.1365-2915.2006.00657.x.
Kajunguri D, Hargrove JW, Ouifki R, Mugisha JY, Coleman PG, Welburn SC: Modelling the use of insecticide-treated cattle to control tsetse and Trypanosoma brucei rhodesiense in a multi-host population. Bull Math Biol. 2014, 76 (3): 673-696. 10.1007/s11538-014-9938-6.
Acknowledgements
The research leading to these results has received funding from the European Union's Seventh Framework Program (FP7/2007-2013) under grant agreement n° 221948, ICONZ (Integrated Control of Neglected Zoonoses) to CW and JDK and Carnegie-Makerere University Next Generation of African Academics under grant number NGAA-2010-2012 to MD. Authors wish to acknowledge the assistance of Tororo District Veterinary officer; Dr. Tegule Mukonge and that of the other Tororo District Department of Production staff for the assistance they offered during collection of cattle blood samples. We also acknowledge Ward Bryssinckx of Avia-GIS, Belgium for helping with use of ArcMap software in general mapping and spatial data analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors hereby declare no competing interests. The sponsors had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors’ contributions
DM, KP, JH, MT, SCW, JDK and CW conceived and designed this study. DM carried out blood sample collection and PCR analyses while DM and JH carried out statistical analyses. All authors participated in the manuscript write-up, read the final version and approved it to be submitted for publication.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Muhanguzi, D., Picozzi, K., Hattendorf, J. et al. The burden and spatial distribution of bovine African trypanosomes in small holder crop-livestock production systems in Tororo District, south-eastern Uganda. Parasites Vectors 7, 603 (2014). https://doi.org/10.1186/s13071-014-0603-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-014-0603-6